5JWP
 
 | | Crystal structure of human FIH D201E variant in complex with Zn, alpha-ketoglutarate, and HIF1 alpha peptide. | | Descriptor: | 2-OXOGLUTARIC ACID, GLYCEROL, Hypoxia-inducible factor 1-alpha, ... | | Authors: | Taabazuing, C.Y, Garman, S.C, Eron, S, Knapp, M.J. | | Deposit date: | 2016-05-12 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The facial triad in the alpha-ketoglutarate dependent oxygenase FIH: A role for sterics in linking substrate binding to O2 activation.
J.Inorg.Biochem., 166, 2016
|
|
6Z6F
 
 | | HDAC-PC | | Descriptor: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
6Z6H
 
 | | HDAC-DC | | Descriptor: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (8.55 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
8TWA
 
 | | Cryo-EM structure of S. cerevisiae Ctf18-RFC-PCNA-PolE-DNA complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18, Chromosome transmission fidelity protein 8, ... | | Authors: | Yuan, Z, Georgescu, R, O'Donnell, M, Li, H. | | Deposit date: | 2023-08-20 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Mechanism of PCNA loading by Ctf18-RFC for leading-strand DNA synthesis.
Science, 385, 2024
|
|
6Z6O
 
 | | HDAC-TC | | Descriptor: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
4ZF7
 
 | | Crystal structure of ferret interleukin-2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Interleukin 2, PENTAETHYLENE GLYCOL, ... | | Authors: | Ren, B, Newman, J, McKinstry, W.J, Adams, T.E. | | Deposit date: | 2015-04-21 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Structural and functional characterisation of ferret interleukin-2.
Dev.Comp.Immunol., 55, 2015
|
|
8JRV
 
 | | Cryo-EM structure of the glucagon receptor bound to glucagon and beta-arrestin 1 | | Descriptor: | Beta-arrestin 1 and single-chain fragment variable 30 (scFv30), Glucagon, HA signal peptide,HPC4 purification tag,Glucagon receptor,C-terminal tail of Vasopressin V2 receptor, ... | | Authors: | Chen, K, Zhang, C, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Tail engagement of arrestin at the glucagon receptor.
Nature, 620, 2023
|
|
5HRF
 
 | |
5HRL
 
 | |
1WLJ
 
 | | human ISG20 | | Descriptor: | ACETATE ION, MANGANESE (II) ION, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Horio, T, Murai, M, Inoue, T, Hamasaki, T, Tanaka, T, Ohgi, T. | | Deposit date: | 2004-06-28 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human ISG20, an interferon-induced antiviral ribonuclease
Febs Lett., 577, 2004
|
|
4WWA
 
 | | Crystal structure of binary complex Bud32-Cgi121 | | Descriptor: | EKC/KEOPS complex subunit BUD32, EKC/KEOPS complex subunit CGI121, SULFATE ION | | Authors: | Zhang, W, van Tilbeurgh, H. | | Deposit date: | 2014-11-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.953 Å) | | Cite: | Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex.
Nucleic Acids Res., 43, 2015
|
|
4WW7
 
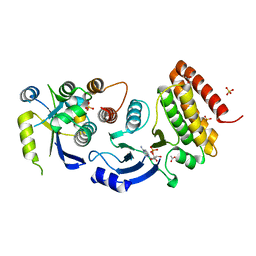 | |
4WX8
 
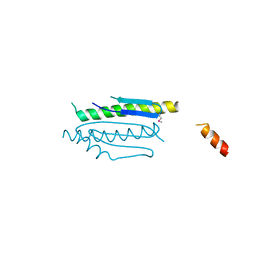 | | Crystal structure of binary complex Gon7-Pcc1 | | Descriptor: | ACETATE ION, EKC/KEOPS complex subunit GON7, EKC/KEOPS complex subunit PCC1 | | Authors: | Zhang, W, Van Tilbeurgh, H. | | Deposit date: | 2014-11-13 | | Release date: | 2015-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex.
Nucleic Acids Res., 43, 2015
|
|
4WXA
 
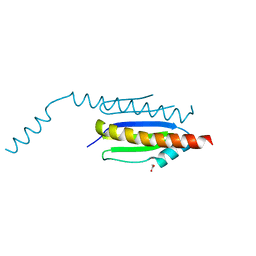 | | Crystal structure of binary complex Gon7-Pcc1 | | Descriptor: | ACETATE ION, EKC/KEOPS complex subunit GON7, EKC/KEOPS complex subunit PCC1, ... | | Authors: | Zhang, W, van Tilbeurgh, H. | | Deposit date: | 2014-11-13 | | Release date: | 2015-03-18 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex.
Nucleic Acids Res., 43, 2015
|
|
4WW5
 
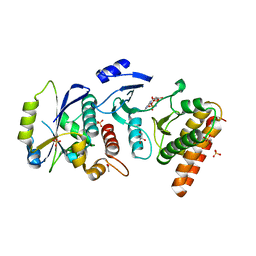 | | Crystal structure of binary complex Bud32-Cgi121 in complex with AMPP | | Descriptor: | ACETATE ION, EKC/KEOPS complex subunit BUD32, EKC/KEOPS complex subunit CGI121, ... | | Authors: | Zhang, W. | | Deposit date: | 2014-11-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex.
Nucleic Acids Res., 43, 2015
|
|
4WW9
 
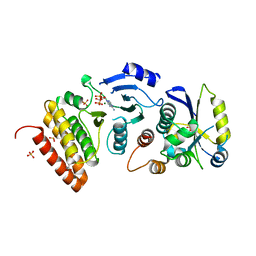 | |
8YBK
 
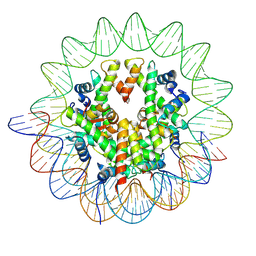 | | Cryo-EM structure of the human nucleosome containing the H3.1 E97K mutant | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kimura, T, Hirai, S, Kujirai, T, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2024-02-14 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Cryo-EM structure and biochemical analyses of the nucleosome containing the cancer-associated histone H3 mutation E97K.
Genes Cells, 29, 2024
|
|
8YBJ
 
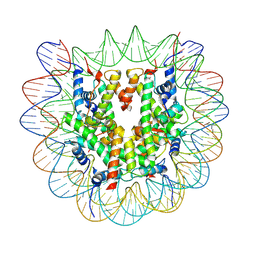 | | Cryo-EM structure of human nucleosome core particle composed of the Widom 601 DNA sequence | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kimura, T, Hirai, S, Kujirai, T, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2024-02-14 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Cryo-EM structure and biochemical analyses of the nucleosome containing the cancer-associated histone H3 mutation E97K.
Genes Cells, 29, 2024
|
|
7LUV
 
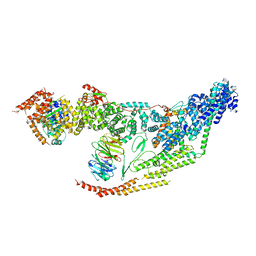 | | Cryo-EM structure of the yeast THO-Sub2 complex | | Descriptor: | ATP-dependent RNA helicase SUB2, THO complex subunit 2, THO complex subunit HPR1, ... | | Authors: | Xie, Y, Ren, Y. | | Deposit date: | 2021-02-23 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the yeast TREX complex and coordination with the SR-like protein Gbp2.
Elife, 10, 2021
|
|
5HRH
 
 | |
5HRK
 
 | |
6JWP
 
 | | crystal structure of EGOC | | Descriptor: | Ego2, GTP-binding protein GTR1, GTP-binding protein GTR2, ... | | Authors: | Zhang, T, Ding, J. | | Deposit date: | 2019-04-21 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the EGO-TC-mediated membrane tethering of the TORC1-regulatory Rag GTPases.
Sci Adv, 5, 2019
|
|
5BNJ
 
 | | CDK8/CYCC IN COMPLEX WITH 8-{3-Chloro-5-[4-(1-methyl-1H-pyrazol-4-yl)-phenyl]-pyridin- 4-yl}-2,8-diaza-spiro[4.5]decan-1-one | | Descriptor: | 8-{3-chloro-5-[4-(1-methyl-1H-pyrazol-4-yl)phenyl]pyridin-4-yl}-2,8-diazaspiro[4.5]decan-1-one, Cyclin-C, Cyclin-dependent kinase 8, ... | | Authors: | Musil, D, Blagg, J, Wienke, D. | | Deposit date: | 2015-05-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | A selective chemical probe for exploring the role of CDK8 and CDK19 in human disease.
Nat.Chem.Biol., 11, 2015
|
|
7K3J
 
 | |
7K3K
 
 | |
