6NTU
 
 | |
4HSJ
 
 | | 1.88 angstrom x-ray crystal structure of piconlinic-bound 3-hydroxyanthranilate-3,4-dioxygenase | | Descriptor: | 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION, PYRIDINE-2-CARBOXYLIC ACID | | Authors: | Liu, F, Chen, L, Davis, C.I, Liu, A. | | Deposit date: | 2012-10-30 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.883 Å) | | Cite: | An Iron Reservoir to the Catalytic Metal: THE RUBREDOXIN IRON IN AN EXTRADIOL DIOXYGENASE.
J.Biol.Chem., 290, 2015
|
|
5OMX
 
 | |
5KSI
 
 | | Crystal structure of deoxygenated hemoglobin in complex with sphingosine phosphate and 2,3-Bisphosphoglycerate | | Descriptor: | (2R)-2,3-diphosphoglyceric acid, (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Hemoglobin subunit alpha, ... | | Authors: | Ahmed, M.H, Safo, M.K, Xia, Y. | | Deposit date: | 2016-07-08 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Insight of Sphingosine 1-Phosphate-Mediated Pathogenic Metabolic Reprogramming in Sickle Cell Disease.
Sci Rep, 7, 2017
|
|
4HUF
 
 | | Structure of 5-chlorouracil modified A:U base pair | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*CP*GP*AP*AP*(UCL)P*TP*CP*GP*CP*G)-3'), GLYCEROL, ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2012-11-02 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure, stability and function of 5-chlorouracil modified A:U and G:U base pairs.
Nucleic Acids Res., 41, 2013
|
|
5AE4
 
 | | Structures of inactive and activated DntR provide conclusive evidence for the mechanism of action of LysR transcription factors | | Descriptor: | LYSR-TYPE REGULATORY PROTEIN, THIOCYANATE ION | | Authors: | Lerche, M, Dian, C, Round, A, Lonneborg, R, Brzezinski, P, Leonard, G.A. | | Deposit date: | 2015-08-25 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Solution Configurations of Inactive and Activated Dntr Have Implications for the Sliding Dimer Mechanism of Lysr Transcription Factors.
Sci.Rep., 6, 2016
|
|
6NMJ
 
 | | Crystal Structure of Rat Ric-8A G alpha binding domain, "Paratone-N Immersed" | | Descriptor: | Resistance to inhibitors of cholinesterase 8 homolog A (C. elegans) | | Authors: | Zeng, B, Mou, T.C, Sprang, S.R. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure, Function, and Dynamics of the G alpha Binding Domain of Ric-8A.
Structure, 27, 2019
|
|
7ZSX
 
 | |
5F64
 
 | | Putative positive transcription regulator (sensor EvgS) from Shigella flexneri | | Descriptor: | Positive transcription regulator EvgA | | Authors: | Nocek, B, Osipiuk, J, Mulligan, R, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-05 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Putative positive transcription regulator (sensor EvgS) from Shigella flexneri.
to be published
|
|
4HWW
 
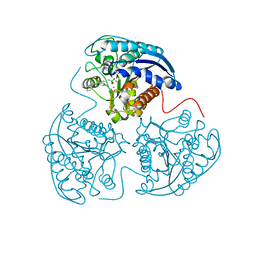 | | Crystal structure of human Arginase-1 complexed with inhibitor 9 | | Descriptor: | Arginase-1, MANGANESE (II) ION, [(5R)-5-amino-5-carboxy-7-(piperidin-1-yl)heptyl](trihydroxy)borate(1-) | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Whitehouse, D.L, Golebiowski, A, Ji, M, Zhang, M, Beckett, P, Sheeler, R, Andreoli, M, Conway, B, Mahboubi, K, Schroeter, H, Van Zandt, M.C, Podjarny, A. | | Deposit date: | 2012-11-09 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | Discovery of (R)-2-Amino-6-borono-2-(2-(piperidin-1-yl)ethyl)hexanoic Acid and Congeners As Highly Potent Inhibitors of Human Arginases I and II for Treatment of Myocardial Reperfusion Injury.
J.Med.Chem., 56, 2013
|
|
5A7X
 
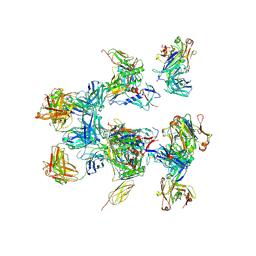 | | negative stain EM of BG505 SOSIP.664 in complex with sCD4, 17b, and 8ANC195 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB OF BROADLY NEUTRALIZING ANTIBODY 17B, FAB OF BROADLY NEUTRALIZING ANTIBODY 8ANC195, ... | | Authors: | Scharf, L, Wang, H, Gao, H, Chen, S, McDowall, A, Bjorkman, P. | | Deposit date: | 2015-07-10 | | Release date: | 2015-08-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Broadly Neutralizing Antibody 8Anc195 Recognizes Closed and Open States of HIV-1 Env.
Cell(Cambridge,Mass.), 162, 2015
|
|
5L1V
 
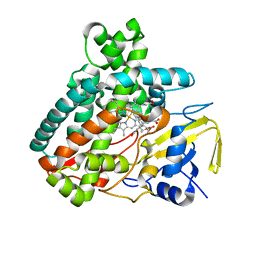 | |
4HX8
 
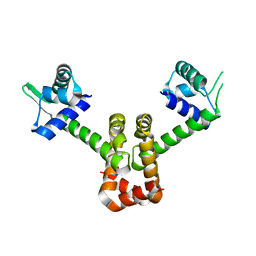 | | Structure of metal-free MNTR mutant E11K | | Descriptor: | Transcriptional regulator MntR | | Authors: | Glasfeld, A, Mcguire, A. | | Deposit date: | 2012-11-09 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Roles of the A and C Sites in the Manganese-Specific Activation of MntR.
Biochemistry, 52, 2013
|
|
5F29
 
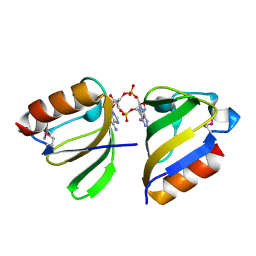 | | Structure of RCK domain with cda | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Na+/H+ antiporter-like protein | | Authors: | Chin, K.H. | | Deposit date: | 2015-12-01 | | Release date: | 2017-02-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | Structural Insights into the Distinct Binding Mode of Cyclic Di-AMP with SaCpaA_RCK.
Biochemistry, 54, 2015
|
|
6KW1
 
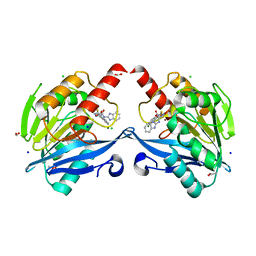 | | The structure of the metallo-beta-lactamase VIM-2 in complex with a triazolylthioacetamide 1b | | Descriptor: | 2-[3-[2-(1H-benzimidazol-2-ylamino)-2-oxidanylidene-ethyl]sulfanyl-1H-1,2,4-triazol-5-yl]benzoic acid, ACETATE ION, Beta-lactamase class B VIM-2, ... | | Authors: | Yang, K.W, Xiang, Y. | | Deposit date: | 2019-09-05 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.775212 Å) | | Cite: | Kinetic, Thermodynamic, and Crystallographic Studies of 2-Triazolylthioacetamides as Verona Integron-Encoded Metallo-beta-Lactamase 2 (VIM-2) Inhibitor.
Biomolecules, 10, 2020
|
|
5IRG
 
 | |
5F2Z
 
 | | Crystal structure of membrane associated PatA from Mycobacterium smegmatis in complex with palmitate - P21 space group | | Descriptor: | PALMITIC ACID, Phosphatidylinositol mannoside acyltransferase | | Authors: | Albesa-Jove, D, Svetlikova, Z, Carreras-Gonzalez, A, Tersa, M, Sancho-Vaello, E, Cifuente, J.O, Mikusova, K, Guerin, M.E. | | Deposit date: | 2015-12-02 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for selective recognition of acyl chains by the membrane-associated acyltransferase PatA.
Nat Commun, 7, 2016
|
|
6KX3
 
 | | Crystal structure of RhoA protein with covalent inhibitor DC-Rhoin | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA, prop-2-enyl (3R)-1,1-bis(oxidanylidene)-2,3-dihydro-1-benzothiophene-3-carboxylate | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2019-09-09 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Covalent Inhibitors Allosterically Block the Activation of Rho Family Proteins and Suppress Cancer Cell Invasion.
Adv Sci, 7, 2020
|
|
5L25
 
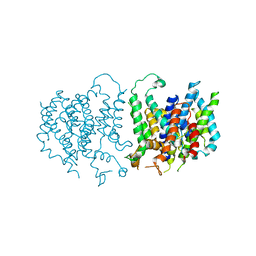 | |
4HXX
 
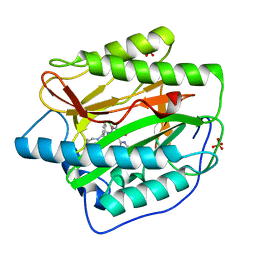 | | Pyridinylpyrimidines selectively inhibit human methionine aminopeptidase-1 | | Descriptor: | (1R)-N~2~-[5-chloro-2-(5-chloropyridin-2-yl)-6-methylpyrimidin-4-yl]-1-phenyl-N~1~-(4-phenylbutyl)ethane-1,2-diamine, COBALT (II) ION, Methionine aminopeptidase 1, ... | | Authors: | Gabelli, S.B, Zhang, F, Liu, J, Amzel, L.M. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Pyridinylpyrimidines selectively inhibit human methionine aminopeptidase-1.
Bioorg.Med.Chem., 21, 2013
|
|
6NN9
 
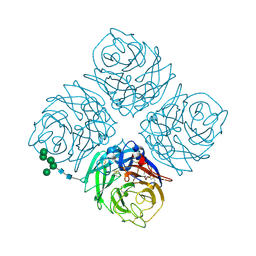 | | REFINED ATOMIC STRUCTURES OF N9 SUBTYPE INFLUENZA VIRUS NEURAMINIDASE AND ESCAPE MUTANTS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE N9, ... | | Authors: | Tulip, W.R, Varghese, J.N, Baker, A.T, Vandonkelaar, A, Laver, W.G, Webster, R.G, Colman, P.M. | | Deposit date: | 1991-03-28 | | Release date: | 1992-07-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined atomic structures of N9 subtype influenza virus neuraminidase and escape mutants.
J.Mol.Biol., 221, 1991
|
|
6L07
 
 | |
7VKT
 
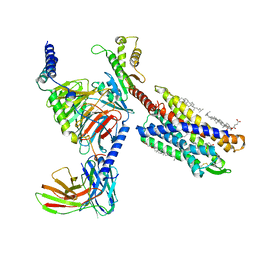 | | cryo-EM structure of LTB4-bound BLT1 in complex with Gi protein | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Wang, N. | | Deposit date: | 2021-10-01 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of leukotriene B4 receptor 1 activation.
Nat Commun, 13, 2022
|
|
7ZRX
 
 | |
6NY0
 
 | |
