5WHK
 
 | | Crystal structure of Fab fragment of antibody DX-2507 bound to FcRn-B2M | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Beta-2-microglobulin, ... | | Authors: | Edwards, T.E, Clifton, M.C, Nixon, A.E, Kenniston, J.A. | | Deposit date: | 2017-07-17 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for pH-insensitive inhibition of immunoglobulin G recycling by an anti-neonatal Fc receptor antibody.
J. Biol. Chem., 292, 2017
|
|
5W7X
 
 | |
5WMI
 
 | | Arabidopsis thaliana Prephenate Aminotransferase mutant- T84V | | Descriptor: | 2-OXOGLUTARIC ACID, Bifunctional aspartate aminotransferase and glutamate/aspartate-prephenate aminotransferase | | Authors: | Jez, J.M, Holland, C.K. | | Deposit date: | 2017-07-28 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for substrate recognition and inhibition of prephenate aminotransferase from Arabidopsis.
Plant J., 94, 2018
|
|
2XGM
 
 | | Substrate and product analogues as human O-GlcNAc transferase inhibitors. | | Descriptor: | ALLOXAN, XCOGT | | Authors: | Dorfmueller, H.C, Borodkin, V.S, Blair, D.E, Pathak, S, Navratilova, I, van Aalten, D.M. | | Deposit date: | 2010-06-07 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Substrate and Product Analogues as Human O-Glcnac Transferase Inhibitors.
Amino Acids, 40, 2011
|
|
1V0O
 
 | | Structure of P. falciparum PfPK5-Indirubin-5-sulphonate ligand complex | | Descriptor: | 2',3-DIOXO-1,1',2',3-TETRAHYDRO-2,3'-BIINDOLE-5'-SULFONIC ACID, CELL DIVISION CONTROL PROTEIN 2 HOMOLOG | | Authors: | Holton, S, Merckx, A, Burgess, D, Doerig, C, Noble, M, Endicott, J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-04-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of P. Falciparum Pfpk5 Test the Cdk Regulation Paradigm and Suggest Mechanisms of Small Molecule Inhibition
Structure, 11, 2003
|
|
4OSQ
 
 | | Crystal structure of the S505R mutant of TAL effector dHax3 | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*TP*AP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*TP*GP*TP*CP*TP*CP*TP*CP*T)-3'), Hax3, ... | | Authors: | Deng, D, Wu, J.P, Yan, C.Y, Pan, X.J, Yan, N. | | Deposit date: | 2014-02-13 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.256 Å) | | Cite: | Revisiting the TALE repeat
Protein Cell, 5, 2014
|
|
1UO4
 
 | | Structure Based Engineering of Internal Molecular Surfaces Of Four Helix Bundles | | Descriptor: | CHLORIDE ION, GENERAL CONTROL PROTEIN GCN4, iodobenzene | | Authors: | Yadav, M.K, Redman, J.E, Alvarez-Gutierrez, J.M, Zhang, Y, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2003-09-15 | | Release date: | 2004-10-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
2XK9
 
 | | Structural analysis of checkpoint kinase 2 (Chk2) in complex with inhibitor PV1533 | | Descriptor: | CHECKPOINT KINASE 2, N-{4-[(1E)-N-(N-hydroxycarbamimidoyl)ethanehydrazonoyl]phenyl}-7-nitro-1H-indole-2-carboxamide | | Authors: | Lountos, G.T, Jobson, A.G, Tropea, J.E, Self, C, Shoemaker, R.H, Pommier, Y, Waugh, D.S. | | Deposit date: | 2010-07-07 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Characterization of Inhibitor Complexes with Checkpoint Kinase 2 (Chk2), a Drug Target for Cancer Therapy.
J.Struct.Biol., 176, 2011
|
|
1ZHI
 
 | | Complex of the S. cerevisiae Orc1 and Sir1 interacting domains | | Descriptor: | Origin recognition complex subunit 1, Regulatory protein SIR1 | | Authors: | Hou, Z, Bernstein, D.A, Fox, C.A, Keck, J.L. | | Deposit date: | 2005-04-25 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the Sir1-origin recognition complex interaction in transcriptional silencing.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
4OVS
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM SULFUROSPIRILLUM DELEYIANUM DSM 6946 (Sdel_0447), TARGET EFI-510309, WITH BOUND SUCCINATE | | Descriptor: | CHLORIDE ION, SUCCINIC ACID, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-13 | | Release date: | 2014-01-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4HZ2
 
 | | Crystal structure of glutathione s-transferase xaut_3756 (target efi-507152) from xanthobacter autotrophicus py2 | | Descriptor: | BENZOIC ACID, GLUTATHIONE, Glutathione S-transferase domain, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N.F, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-11-14 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of glutathione s-transferase xaut_3756 from xanthobacter autotrophicus py2
To be Published
|
|
4HK3
 
 | | I2 Fab (unbound) from CH65-CH67 Lineage | | Descriptor: | I2 heavy chain, I2 light chain | | Authors: | Schmidt, A.G, Harrison, S.C. | | Deposit date: | 2012-10-14 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1UZ0
 
 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with Glc-4Glc-3Glc-4Glc | | Descriptor: | CALCIUM ION, CELLULASE B, CHLORIDE ION, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
3VFT
 
 | | crystal structure of HLA B*3508LPEP-P6Ala, peptide mutant P6-ala | | Descriptor: | Beta-2-microglobulin, LPEP peptide from EBV, P6A, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.947 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
5SXX
 
 | |
4O8H
 
 | | 0.85A resolution structure of PEG 400 Bound Cyclophilin D | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Lovell, S, Valasani, K.R, Battaile, K.P, Wang, C, Yan, S.S. | | Deposit date: | 2013-12-27 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | High-resolution crystal structures of two crystal forms of human cyclophilin D in complex with PEG 400 molecules.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4OZ0
 
 | |
5SXR
 
 | | Crystal structure of B. pseudomallei KatG with NAD bound | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2016-08-10 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Isonicotinic acid hydrazide conversion to Isonicotinyl-NAD by catalase-peroxidases.
J. Biol. Chem., 285, 2010
|
|
2XPL
 
 | | Crystal structure of Iws1(Spn1) conserved domain from Encephalitozoon cuniculi | | Descriptor: | CHLORIDE ION, IWS1 | | Authors: | Koch, M, Diebold, M.-L, Cura, V, Cavarelli, J, Romier, C. | | Deposit date: | 2010-08-27 | | Release date: | 2010-11-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Structure of an Iws1/Spt6 Complex Reveals an Interaction Domain Conserved in Tfiis, Elongin a and Med26
Embo J., 29, 2010
|
|
2XGB
 
 | | Crystal structure of Barley Beta-Amylase complexed with 2,3- epoxypropyl-alpha-D-glucopyranoside | | Descriptor: | (2R)-oxiran-2-ylmethyl alpha-D-glucopyranoside, 1,2-ETHANEDIOL, BETA-AMYLASE | | Authors: | Rejzek, M, Stevenson, C.E.M, Southard, A.M, Stanley, D, Denyer, K, Smith, A.M, Naldrett, M.J, Lawson, D.M, Field, R.A. | | Deposit date: | 2010-06-02 | | Release date: | 2010-12-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Chemical Genetics and Cereal Starch Metabolism: Structural Basis of the Non-Covalent and Covalent Inhibition of Barley Beta-Amylase.
Mol.Biosyst., 7, 2011
|
|
3VPN
 
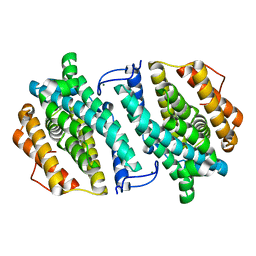 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | Descriptor: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | Authors: | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Essential role of E106 in the proton-coupled electron transfer in human
to be published
|
|
4L0M
 
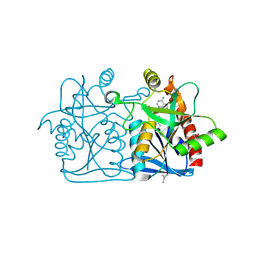 | |
4OCA
 
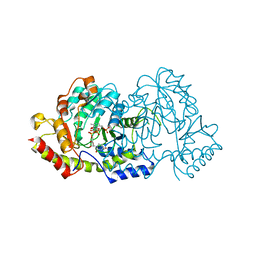 | | Cryatal structure of ArnB K188A complexted with PLP and UDP-Ara4N | | Descriptor: | (2R,3R,4S,5S)-3,4-dihydroxy-5-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]tetrahydro-2H-pyr an-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, UDP-4-amino-4-deoxy-L-arabinose--oxoglutarate aminotransferase | | Authors: | Sousa, M.C, Lee, M. | | Deposit date: | 2014-01-08 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Substrate Specificity in ArnB. A Key Enzyme in the Polymyxin Resistance Pathway of Gram-Negative Bacteria.
Biochemistry, 53, 2014
|
|
4ODP
 
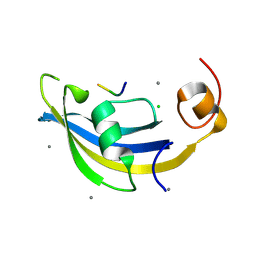 | | Structure of SlyD delta-IF from Thermus thermophilus in complex with S2-W23A peptide | | Descriptor: | 30S ribosomal protein S2, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Quistgaard, E.M, Low, C, Nordlund, P. | | Deposit date: | 2014-01-10 | | Release date: | 2015-01-14 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Molecular insights into substrate recognition and catalytic mechanism of the chaperone and FKBP peptidyl-prolyl isomerase SlyD.
BMC Biol., 14, 2016
|
|
4OC6
 
 | | Structure of Cathepsin D with inhibitor 2-bromo-N-[(2S,3S)-4-{[2-(2,4-dichlorophenyl)ethyl][3-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)propanoyl]amino}-3-hydroxy-1-(3-phenoxyphenyl)butan-2-yl]-4,5-dimethoxybenzamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-bromo-N-[(2S,3S)-4-{[2-(2,4-dichlorophenyl)ethyl][3-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)propanoyl]amino}-3-hydroxy-1-(3-phenoxyphenyl)butan-2-yl]-4,5-dimethoxybenzamide, Cathepsin D heavy chain, ... | | Authors: | Graedler, U, Czodrowski, P, Tsaklakidis, C, Klein, M, Maskos, K, Leuthner, B. | | Deposit date: | 2014-01-08 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure-based optimization of non-peptidic Cathepsin D inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
