6BK0
 
 | |
7DZX
 
 | | Spike protein from SARS-CoV2 with Fab fragment of enhancing antibody 8D2 | | Descriptor: | Fab Heavy chain of enhancing antibody, Fab light chain of enhancing antibody, Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
7W0A
 
 | | dmDicer2-LoqsPD-dsRNA Dimer status | | Descriptor: | Dicer-2, isoform A, Loquacious, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
6BKD
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 6a bound to broadly neutralizing antibody AR3D | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab AR3D heavy chain, Fab AR3D light chain, ... | | Authors: | Tzarum, N, Wilson, I.A, Law, M. | | Deposit date: | 2017-11-08 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Genetic and structural insights into broad neutralization of hepatitis C virus by human VH1-69 antibodies.
Sci Adv, 5, 2019
|
|
6TDM
 
 | | Bam_5920cDD 5919nDD docking domains | | Descriptor: | Beta-ketoacyl synthase,Beta-ketoacyl synthase | | Authors: | Risser, F, Chagot, B. | | Deposit date: | 2019-11-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: Docking in the enacyloxin IIa polyketide synthase.
J.Struct.Biol., 212, 2020
|
|
6TRM
 
 | | Solution structure of the antifungal protein PAFC | | Descriptor: | Pc21g12970 protein | | Authors: | Czajlik, A, Holzknecht, J, Marx, F, Batta, G. | | Deposit date: | 2019-12-19 | | Release date: | 2020-10-28 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Dynamics, and New Antifungal Aspects of the Cysteine-Rich Miniprotein PAFC.
Int J Mol Sci, 22, 2021
|
|
8HMY
 
 | | Cryo-EM structure of the human pre-catalytic TSEN/pre-tRNA complex | | Descriptor: | Chromosome 1 open reading frame 19, isoform CRA_a, MAGNESIUM ION, ... | | Authors: | Zhang, X, Yang, F, Zhan, X, Shi, Y. | | Deposit date: | 2022-12-06 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural basis of pre-tRNA intron removal by human tRNA splicing endonuclease.
Mol.Cell, 83, 2023
|
|
7W0E
 
 | | dmDicer2-LoqsPD-dsRNA Active-dicing status | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dicer-2, isoform A, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
5N6N
 
 | | CRYSTAL STRUCTURE OF THE 14-3-3:NEUTRAL TREHALASE NTH1 COMPLEX | | Descriptor: | CALCIUM ION, Neutral trehalase, Protein BMH1, ... | | Authors: | Alblova, M, Smidova, A, Obsilova, V, Obsil, T. | | Deposit date: | 2017-02-15 | | Release date: | 2017-11-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Molecular basis of the 14-3-3 protein-dependent activation of yeast neutral trehalase Nth1.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7W0C
 
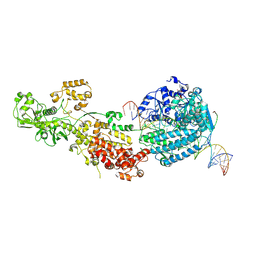 | | Dicer2-Loqs-PD-dsRNA complex at early-translocation state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dicer-2, isoform A, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
5N0I
 
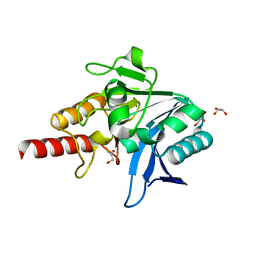 | | Crystal structure of NDM-1 in complex with beta-mercaptoethanol - new refinement | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, King, D.T, Strynadka, N.C.J. | | Deposit date: | 2017-02-03 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
7W0B
 
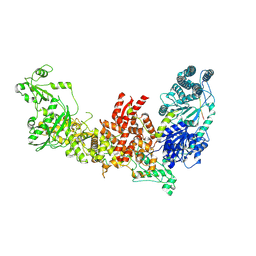 | | Dicer2-LoqsPD complex at apo status | | Descriptor: | Dicer-2, isoform A, Loquacious, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
6TDN
 
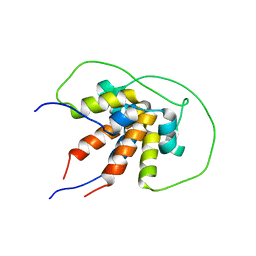 | | Bam_5925cDD 5924nDD docking domains | | Descriptor: | Beta-ketoacyl synthase,Beta-ketoacyl synthase | | Authors: | Risser, F, Chagot, B. | | Deposit date: | 2019-11-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: Docking in the enacyloxin IIa polyketide synthase.
J.Struct.Biol., 212, 2020
|
|
7W0F
 
 | | dmDicer2-LoqsPD-dsRNA Post-dicing status | | Descriptor: | Dicer-2, isoform A, Loquacious, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
6QN6
 
 | |
8HO0
 
 | | Crystal structure of cytochrome P450 NasF5053 mutant E73S complexed with 8FCWP | | Descriptor: | (3~{S},8~{a}~{S})-3-[(7-fluoranyl-1~{H}-indol-3-yl)methyl]-2,3,6,7,8,8~{a}-hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, CALCIUM ION, Cytochrome P450-F5053, ... | | Authors: | Ma, B.D, Tian, W, Qu, X, Kong, X.D. | | Deposit date: | 2022-12-09 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Engineering the Substrate Specificity of a P450 Dimerase Enables the Collective Biosynthesis of Heterodimeric Tryptophan-Containing Diketopiperazines.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6NLB
 
 | | Crystal structure of de novo designed metal-controlled dimer of mutant B1 immunoglobulin-binding domain of Streptococcal Protein G (L12H, E15V, T16L, T18I, V29H, Y33H, N37L)-apo | | Descriptor: | Immunoglobulin G-binding protein G, MAGNESIUM ION | | Authors: | Maniaci, B, Stec, B, Huxford, T. | | Deposit date: | 2019-01-08 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of High-Affinity Metal-Controlled Protein Dimers.
Biochemistry, 58, 2019
|
|
5BSG
 
 | | Crystal structure of Medicago truncatula (delta)1-Pyrroline-5-Carboxylate Reductase (MtP5CR) in complex with NADP+ | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Ruszkowski, M, Nocek, B, Forlani, G, Dauter, Z. | | Deposit date: | 2015-06-02 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of Medicago truncatula delta (1)-pyrroline-5-carboxylate reductase provides new insights into regulation of proline biosynthesis in plants.
Front Plant Sci, 6, 2015
|
|
6NLL
 
 | | 1.80 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor (analog 14) | | Descriptor: | 4-{[3-(2,4-dihydroxyphenyl)propyl]amino}-1H-isoindole-1,3(2H)-dione, FE (II) ION, Ferroxidase, ... | | Authors: | Lovell, S, Punchi-Hewage, A, Battaile, K.P, Yao, H, Nammalwar, B, Gnanasekaran, K.K, Bunce, R.A, Reitz, A.B, Rivera, M. | | Deposit date: | 2019-01-08 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small Molecule Inhibitors of the BfrB-Bfd Interaction Decrease Pseudomonas aeruginosa Fitness and Potentiate Fluoroquinolone Activity.
J.Am.Chem.Soc., 141, 2019
|
|
6BPG
 
 | |
1OUB
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: X-RAY STRUCTURE OF THE V100A MUTANT | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of the hydrophobic effect to the stability of human lysozyme: calorimetric studies and X-ray structural analyses of the nine valine to alanine mutants.
Biochemistry, 36, 1997
|
|
6TDD
 
 | | Bam_5924 docking domain | | Descriptor: | Beta-ketoacyl synthase | | Authors: | Risser, F, Chagot, B. | | Deposit date: | 2019-11-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: Docking in the enacyloxin IIa polyketide synthase.
J.Struct.Biol., 212, 2020
|
|
1OUH
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: X-RAY STRUCTURE OF THE V74A MUTANT | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of the hydrophobic effect to the stability of human lysozyme: calorimetric studies and X-ray structural analyses of the nine valine to alanine mutants.
Biochemistry, 36, 1997
|
|
2LWQ
 
 | |
7XX3
 
 | | Crystal structure of human Superoxide Dismutase (SOD1) in complex with a fungal metabolite molecule, Phialomustin B (PB) | | Descriptor: | (2~{E},4~{E},6~{S})-4,6-dimethyldeca-2,4-dienoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Padmanabhan, B, Unni, S. | | Deposit date: | 2022-05-28 | | Release date: | 2023-12-13 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the modulation Of SOD1 aggregation By a fungal metabolite Phialomustin-B: Therapeutic potential in ALS.
Plos One, 19, 2024
|
|
