4PEY
 
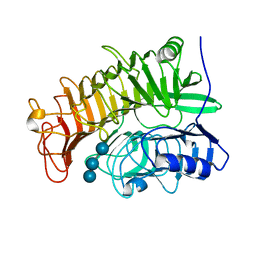 | | Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritriose | | Descriptor: | Putative secreted protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-04-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
4PEZ
 
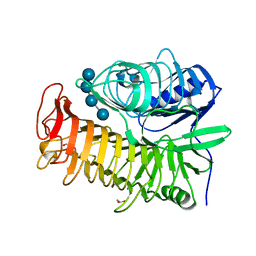 | | Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritetraose | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted protein, beta-D-glucopyranose, ... | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-04-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
3IOQ
 
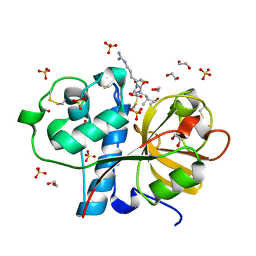 | | Crystal structure of the Carica candamarcensis cysteine protease CMS1MS2 in complex with E-64. | | Descriptor: | 1,2-ETHANEDIOL, CMS1MS2, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, ... | | Authors: | Gomes, M.T.R, Teixeira, R.D, Salas, C.E, Nagem, R.A.P. | | Deposit date: | 2009-08-14 | | Release date: | 2010-02-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of the Carica candamarcensis cysteine protease CMS1MS2 in complex with E-64
To be Published
|
|
1ZMR
 
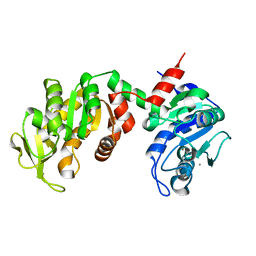 | | Crystal Structure of the E. coli Phosphoglycerate Kinase | | Descriptor: | CALCIUM ION, Phosphoglycerate kinase | | Authors: | Marqusee, S. | | Deposit date: | 2005-05-10 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comparison of proteolytic susceptibility in phosphoglycerate kinases from yeast and E. coli: modulation of conformational ensembles without altering structure or stability.
J.Mol.Biol., 368, 2007
|
|
1LEL
 
 | | The avidin BCAP complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Avidin, E-AMINO BIOTINYL CAPROIC ACID | | Authors: | Pazy, Y, Kulik, T, Bayer, E.A, Wilchek, M, Livnah, O. | | Deposit date: | 2002-04-10 | | Release date: | 2002-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand exchange between proteins: exchange of biotin and biotin derivatives between avidin and streptavidin
J.Biol.Chem., 277, 2002
|
|
4PF0
 
 | | Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminarihexaose | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-04-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
4ZSY
 
 | | Pig Brain GABA-AT inactivated by (Z)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic acid. | | Descriptor: | (1S)-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopent-3-ene-1,3-dicarboxylic acid, 4-aminobutyrate aminotransferase, mitochondrial, ... | | Authors: | Wu, R, Lee, H, Le, H.V, Doud, E, Sanishvili, R, Compton, P, Kelleher, N.L, Silverman, R.B, Liu, D. | | Deposit date: | 2015-05-14 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Inactivation of GABA Aminotransferase by (E)- and (Z)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic Acid.
Acs Chem.Biol., 10, 2015
|
|
4PEW
 
 | | Structure of sacteLam55A from Streptomyces sp. SirexAA-E | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Putative secreted protein | | Authors: | Bianchetti, C.M, Takasuka, T.E, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-04-25 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
4PEX
 
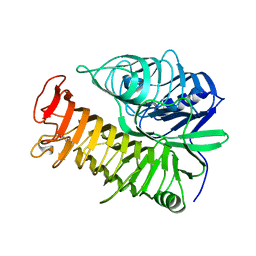 | | Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted protein, beta-D-glucopyranose | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-04-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
4C2I
 
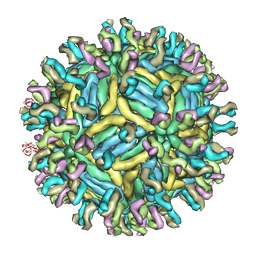 | | Cryo-EM structure of Dengue virus serotype 1 complexed with Fab fragments of human antibody 1F4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fibriansah, G, Tan, J.L, de Alwis, R, Smith, S.A, Ng, T.-S, Kostyuchenko, V.A, Ibarra, K.D, Harris, E, de Silva, A, Crowe Junior, J.E, Lok, S.-M. | | Deposit date: | 2013-08-18 | | Release date: | 2014-01-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | A Potent Anti-Dengue Human Antibody Preferentially Recognizes the Conformation of E Protein Monomers Assembled on the Virus Surface.
Embo Mol.Med., 6, 2014
|
|
5AMT
 
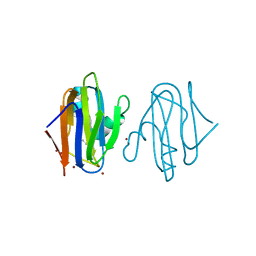 | | Intracellular growth locus protein E | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, IGLE | | Authors: | Robb, C.S, Nano, F.E, Boraston, A.B. | | Deposit date: | 2015-09-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Cloning, Expression, Purification, Crystallization and Preliminary X-Ray Diffraction Analysis of Intracellular Growth Locus E (Igle) Protein from Francisella Tularensis Subsp. Novicida.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
7JQW
 
 | |
1MJZ
 
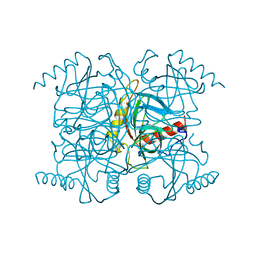 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE MUTANT D97N | | Descriptor: | INORGANIC PYROPHOSPHATASE | | Authors: | Oganesyan, V, Harutyunyan, E.H, Avaeva, S.M, Huber, R. | | Deposit date: | 1997-02-08 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of mutant forms of E. coli inorganic pyrophosphatase with Asp-->Asn single substitution in positions 42, 65, 70, and 97.
Biochemistry Mosc., 63, 1998
|
|
1MJY
 
 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE MUTANT D70N | | Descriptor: | INORGANIC PYROPHOSPHATASE | | Authors: | Oganesyan, V, Harutyunyan, E.H, Avaeva, S.M, Huber, R. | | Deposit date: | 1997-02-08 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structures of mutant forms of E. coli inorganic pyrophosphatase with Asp-->Asn single substitution in positions 42, 65, 70, and 97.
Biochemistry Mosc., 63, 1998
|
|
3GIC
 
 | | Structure of thrombin mutant delta(146-149e) in the free form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Thrombin heavy chain, Thrombin light chain | | Authors: | Bah, A, Carrell, C.J, Chen, Z, Gandhi, P.S, Di Cera, E. | | Deposit date: | 2009-03-05 | | Release date: | 2009-06-02 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Stabilization of the E* form turns thrombin into an anticoagulant.
J.Biol.Chem., 284, 2009
|
|
1JWE
 
 | | NMR Structure of the N-Terminal Domain of E. Coli Dnab Helicase | | Descriptor: | PROTEIN (DNAB HELICASE) | | Authors: | Weigelt, J, Brown, S.E, Miles, C.S, Dixon, N.E, Otting, G. | | Deposit date: | 1999-01-22 | | Release date: | 1999-01-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the N-terminal domain of E. coli DnaB helicase: implications for structure rearrangements in the helicase hexamer.
Structure Fold.Des., 7, 1999
|
|
3JCX
 
 | | Canine Parvovirus complexed with Fab E | | Descriptor: | Capsid protein 2, Fab E heavy chain, Fab E light chain | | Authors: | Organtini, L.J, Iketani, S, Huang, K, Ashley, R.E, Makhov, A.M, Conway, J.F, Parrish, C.R, Hafenstein, S. | | Deposit date: | 2016-03-21 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Near-Atomic Resolution Structure of a Highly Neutralizing Fab Bound to Canine Parvovirus.
J.Virol., 90, 2016
|
|
4TYV
 
 | | Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted protein, beta-D-glucopyranose | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-07-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
2OVR
 
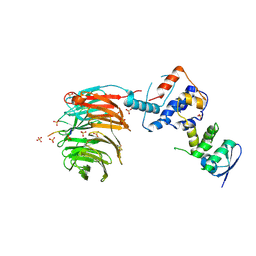 | | Structure of the Skp1-Fbw7-CyclinEdegN complex | | Descriptor: | F-box/WD repeat protein 7, S-phase kinase-associated protein 1A, SULFATE ION, ... | | Authors: | Hao, B, Oehlmann, S, Sowa, M.E, Harper, J.W, Pavletich, N.P. | | Deposit date: | 2007-02-14 | | Release date: | 2007-04-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a Fbw7-Skp1-Cyclin E Complex: Multisite-Phosphorylated Substrate Recognition by SCF Ubiquitin Ligases
Mol.Cell, 26, 2007
|
|
3GLZ
 
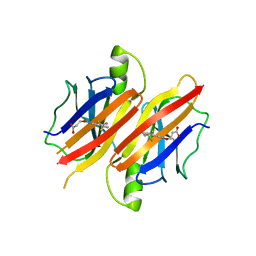 | | Human Transthyretin (TTR) complexed with(E)-3-(2-(trifluoromethyl)benzylideneaminooxy)propanoic acid (inhibitor 11) | | Descriptor: | 3-[({(1E)-[2-(trifluoromethyl)phenyl]methylidene}amino)oxy]propanoic acid, Transthyretin | | Authors: | Mohamedmohaideen, N.N, Palaninathan, S.K, Orlandini, E, Sacchettini, J.C. | | Deposit date: | 2009-03-12 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Novel transthyretin amyloid fibril formation inhibitors: synthesis, biological evaluation, and X-ray structural analysis
Plos One, 4, 2009
|
|
5QIB
 
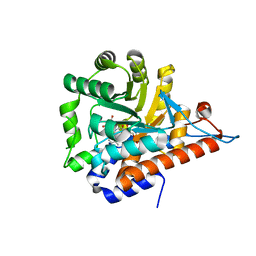 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of HAO1 in complex with FMOPL000388a | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 5-(cyclohexylamino)pyrimidine-2,4(1H,3H)-dione, Hydroxyacid oxidase 1 | | Authors: | MacKinnon, S, Bezerra, G.A, Krojer, T, Bradley, A.R, Talon, R, Brandao-Neto, J, Douangamath, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Brennan, P.E, Yue, W.W. | | Deposit date: | 2018-05-22 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
1YVQ
 
 | | The low salt (PEG) crystal structure of CO Hemoglobin E (betaE26K) approaching physiological pH (pH 7.5) | | Descriptor: | CARBON MONOXIDE, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Malashkevich, V.N, Balazs, T.C, Almo, S.C, Hirsch, R.E. | | Deposit date: | 2005-02-16 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of CO Hemoglobin E (betaE26K) approaching physiological pH (pH 7.5)
To be Published
|
|
2OVP
 
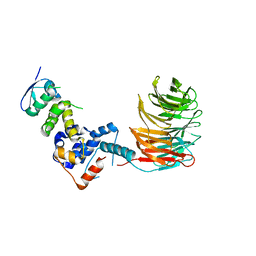 | | Structure of the Skp1-Fbw7 complex | | Descriptor: | F-box/WD repeat protein 7, S-phase kinase-associated protein 1A | | Authors: | Hao, B, Oehlmann, S, Sowa, M.E, Harper, J.W, Pavletich, N.P. | | Deposit date: | 2007-02-14 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a Fbw7-Skp1-Cyclin E Complex: Multisite-Phosphorylated Substrate Recognition by SCF Ubiquitin Ligases
Mol.Cell, 26, 2007
|
|
3ZDB
 
 | | Structure of E. coli ExoIX in complex with the palindromic 5ov4 DNA oligonucleotide, di-magnesium and potassium | | Descriptor: | 5OV4 DNA, 5'-D(*AP*AP*AP*AP*GP*CP*GP*TP*AP*CP*GP*CP)-3', ACETATE ION, ... | | Authors: | Hemsworth, G.R, Anstey-Gilbert, C.S, Flemming, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The Structure of E. Coli Exoix - Implications for DNA Binding and Catalysis in Flap Endonucleases
Nucleic Acids Res., 41, 2013
|
|
3ZDE
 
 | | Potassium free structure of E. coli ExoIX | | Descriptor: | PROTEIN XNI | | Authors: | Hemsworth, G.R, Anstey-Gilbert, C.S, Flemming, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The Structure of E. Coli Exoix - Implications for DNA Binding and Catalysis in Flap Endonucleases
Nucleic Acids Res., 41, 2013
|
|
