9FJY
 
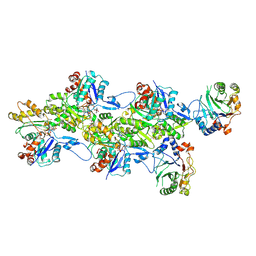 | | Structure of the DNase I- and phalloidin-bound pointed end of F-actin (conformer 2). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Boiero Sanders, M, Oosterheert, W, Hofnagel, O, Bieling, P, Raunser, S. | | Deposit date: | 2024-05-31 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Phalloidin and DNase I-bound F-actin pointed end structures reveal principles of filament stabilization and disassembly.
Nat Commun, 15, 2024
|
|
9FJW
 
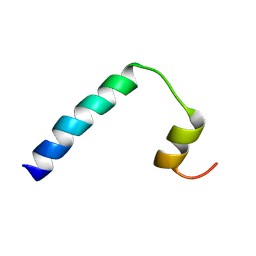 | | Solution NMR structure of a peptide encompassing residues 2-36 of the human formin INF2 | | Descriptor: | Inverted formin-2 | | Authors: | Jimenez, M.A, Comas, L, Labat-de-Hoz, L, Correas, I, Alonso, M.A. | | Deposit date: | 2024-05-31 | | Release date: | 2024-09-11 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the N-terminal extension of the formin INF2.
Cell Mol Life Sci, 79, 2022
|
|
9FJU
 
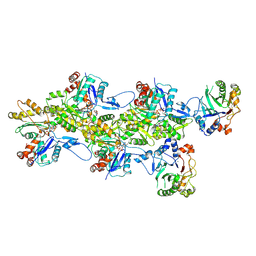 | | Structure of the DNase I- and phalloidin-bound pointed end of F-actin (conformer 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Boiero Sanders, M, Oosterheert, W, Hofnagel, O, Bieling, P, Raunser, S. | | Deposit date: | 2024-05-31 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Phalloidin and DNase I-bound F-actin pointed end structures reveal principles of filament stabilization and disassembly.
Nat Commun, 15, 2024
|
|
9FJO
 
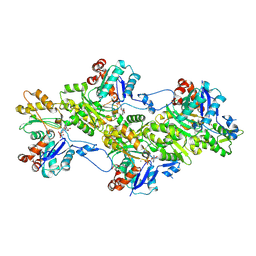 | | Structure of the undecorated pointed end of F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Boiero Sanders, M, Oosterheert, W, Hofnagel, O, Bieling, P, Raunser, S. | | Deposit date: | 2024-05-31 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Phalloidin and DNase I-bound F-actin pointed end structures reveal principles of filament stabilization and disassembly.
Nat Commun, 15, 2024
|
|
9FJN
 
 | | Solution NMR structure of a peptide encompassing residues 2-19 of the human formin INF2 | | Descriptor: | Inverted formin-2 | | Authors: | Jimenez, M.A, Comas, L, Labat-de-Hoz, L, Correas, I, Alonso, M.A. | | Deposit date: | 2024-05-31 | | Release date: | 2024-09-11 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the N-terminal extension of the formin INF2.
Cell Mol Life Sci, 79, 2022
|
|
9FJM
 
 | | Cryo-EM structure of the phalloidin-bound pointed end of the actin filament. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Boiero Sanders, M, Oosterheert, W, Hofnagel, O, Bieling, P, Raunser, S. | | Deposit date: | 2024-05-31 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Phalloidin and DNase I-bound F-actin pointed end structures reveal principles of filament stabilization and disassembly.
Nat Commun, 15, 2024
|
|
9FJL
 
 | |
9FJK
 
 | | Omicron BA.1 Spike protein with neutralizing NTD specific mAb K501SP6 | | Descriptor: | K501SP6 Fv Heavy Chain, K501SP6 Fv Light Chain, Spike glycoprotein,Fibritin | | Authors: | Bjoernsson, K.H, Walker, M.R, Raghavan, S.S.R, Ward, A.B, Barfod, L.K. | | Deposit date: | 2024-05-31 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Omicron BA.1 Spike protein with neutralizing NTD specific mAb K501SP6
To Be Published
|
|
9FJE
 
 | | Expanded formalin inactivated CVB1 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Plavec, Z, Butcher, S.J. | | Deposit date: | 2024-05-31 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Comparison of structure and immunogenicity of CVB1-VLP and inactivated CVB1 vaccine candidates.
Res Sq, 2024
|
|
9FJD
 
 | | Expanded CVB1-VLP (Tween80) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Plavec, Z, Butcher, S.J. | | Deposit date: | 2024-05-31 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Comparison of structure and immunogenicity of CVB1-VLP and inactivated CVB1 vaccine candidates.
Res Sq, 2024
|
|
9FJC
 
 | | Compact CVB1-VLP (Tween80) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Plavec, Z, Butcher, S.J. | | Deposit date: | 2024-05-30 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Comparison of structure and immunogenicity of CVB1-VLP and inactivated CVB1 vaccine candidates.
Res Sq, 2024
|
|
9FJ4
 
 | | Structure of ubiquitin bound to coiled coil-UIM form 1 | | Descriptor: | 3-[1-(2-oxidanylideneethyl)-1,2,3-triazol-4-yl]propanal, GLU-GLN-GLU-ILE-GLU-GLU-LEU-GLU-ILE-GLU-ILE-ALA-ILE-LEU-LEU-SER-GLU-ILE-GLU-GLY, LYS-GLN-LYS-ILE-ALA-ALA-LEU-LYS-TYR-LYS-ILE-ALA-ALA-LEU-LYS-GLN-LYS-ILE-GLN, ... | | Authors: | Paredes Vergara, P, Huang, D.T. | | Deposit date: | 2024-05-30 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | An engineered ubiquitin binding coiled coil peptide.
Chem Sci, 15, 2024
|
|
9FJ3
 
 | | Structure of ubiquitin bound of coiled-coil UIM form 2 | | Descriptor: | 3-[1-(2-oxidanylideneethyl)-1,2,3-triazol-4-yl]propanal, GLU-GLN-GLU-ILE-GLU-GLU-LEU-GLU-ILE-GLU-ILE-ALA-ILE-LEU-LEU-SER-GLU-ILE-GLU-GLY, LYS-GLN-LYS-ILE-ALA-ALA-LEU-LYS-TYR-LYS-ILE-ALA-ALA-LEU-LYS-GLN-LYS-ILE, ... | | Authors: | Paredes Vergara, P, Huang, D.T. | | Deposit date: | 2024-05-30 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An engineered ubiquitin binding coiled coil peptide.
Chem Sci, 15, 2024
|
|
9FIL
 
 | | Crystal Structure of reduced NuoEF variant E222K(NuoF) from Aquifex aeolicus bound to NAD+ | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-29 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FIJ
 
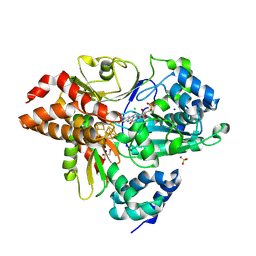 | | Crystal Structure of reduced NuoEF variant E222K(NuoF) from Aquifex aeolicus | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-29 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FII
 
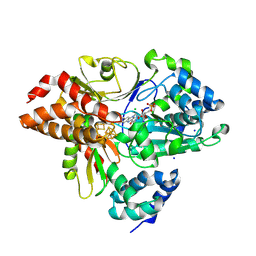 | | Crystal Structure of oxidized NuoEF variant E222K(NuoF) from Aquifex aeolicus | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-29 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FIH
 
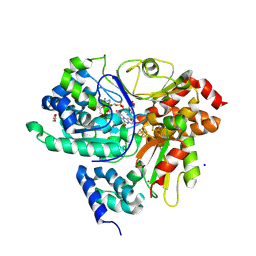 | | Crystal Structure of NuoEF variant P228R(NuoF) from Aquifex aeolicus bound to NADH under anoxic conditions after 10 min soaking | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-29 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FIF
 
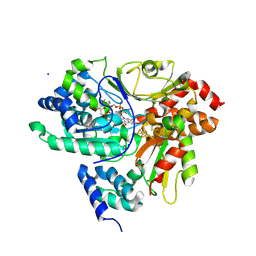 | | Crystal Structure of NuoEF variant P228R(NuoF) from Aquifex aeolicus bound to NADH under anoxic conditions | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-29 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FI9
 
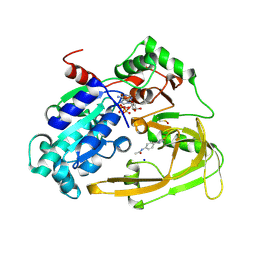 | | Human PIF + Z48847594 (OCCUPANCY 0.7) | | Descriptor: | ATP-dependent DNA helicase PIF1, MAGNESIUM ION, N-[4-(2-amino-1,3-thiazol-4-yl)phenyl]acetamide, ... | | Authors: | Bax, B.D, Sanders, C.M, Antson, A.A, Brandao-Neto, J. | | Deposit date: | 2024-05-28 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.727 Å) | | Cite: | Structure-based discovery of first inhibitors targeting the helicase activity of human PIF1
Nucleic Acids Research, 2024
|
|
9FHP
 
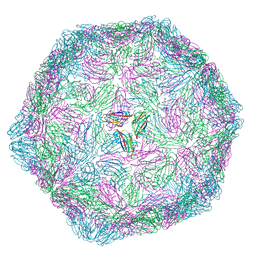 | |
9FHE
 
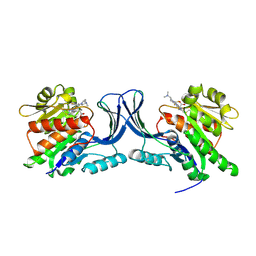 | | hKHK-C in complex with BI-9787 (pH 5.5) | | Descriptor: | (2~{S})-3-[3-[[4-[bis(fluoranyl)methyl]-3-cyano-6-[(3~{S})-3-(dimethylamino)pyrrolidin-1-yl]pyridin-2-yl]amino]-4-methylsulfanyl-phenyl]-2-methyl-propanoic acid, Ketohexokinase | | Authors: | Ebenhoch, R, Pautsch, A. | | Deposit date: | 2024-05-27 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | Discovery of BI-9787, a potent zwitterionic ketohexokinase inhibitor with oral bioavailability.
Bioorg.Med.Chem.Lett., 112, 2024
|
|
9FHD
 
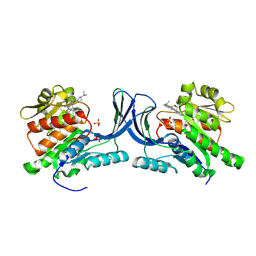 | | hKHK-C in fomplex with BI-9787 | | Descriptor: | (2~{S})-3-[3-[[4-[bis(fluoranyl)methyl]-3-cyano-6-[(3~{S})-3-(dimethylamino)pyrrolidin-1-yl]pyridin-2-yl]amino]-4-methylsulfanyl-phenyl]-2-methyl-propanoic acid, Ketohexokinase, SULFATE ION | | Authors: | Ebenhoch, R, Pautsch, A. | | Deposit date: | 2024-05-27 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Discovery of BI-9787, a potent zwitterionic ketohexokinase inhibitor with oral bioavailability.
Bioorg.Med.Chem.Lett., 112, 2024
|
|
9FH9
 
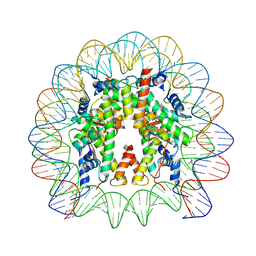 | | Structure of CyclinB1 N-terminus bound to the NCP | | Descriptor: | DNA (145-MER), G2/mitotic-specific cyclin-B1, Histone H2A type 3, ... | | Authors: | Young, R.V.C, Muhammad, R, Alfieri, C. | | Deposit date: | 2024-05-27 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Spatial control of the APC/C ensures the rapid degradation of cyclin B1.
Embo J., 43, 2024
|
|
9FGU
 
 | | SARS-CoV-2 (B.1.1.529/Omicron variant) Spike protein in complex with the single chain fragment scFv76-77 (focused refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, scFv76-77 single chain fragment | | Authors: | Berlinguer, M, Chaves-Sanjuan, A, Milazzo, F.M, Minenkova, O, De Santis, R, Bolognesi, M. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of scFv76-77 in complex with SARS-CoV-2 Omicron Spike protein
To Be Published
|
|
9FGT
 
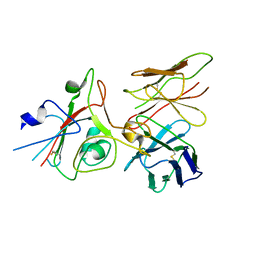 | | SARS-CoV-2 (B.1.1.529/Omicron variant) Spike protein in complex with the single chain fragment scFv76 (focused refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Single chain fragment scFv76, Spike glycoprotein,Fibritin | | Authors: | Berlinguer, M, Chaves-Sanjuan, A, Milazzo, F.M, Minenkova, O, De Santis, R, Bolognesi, M. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of scFv76 in complex with SARS-CoV-2 Omicron Spike protein
To Be Published
|
|
