2HFP
 
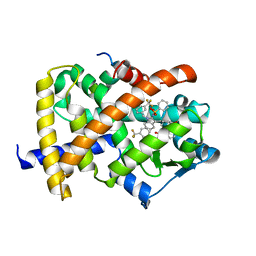 | | Crystal Structure of PPAR Gamma with N-sulfonyl-2-indole carboxamide ligands | | Descriptor: | 3-(4-METHOXYPHENYL)-N-(PHENYLSULFONYL)-1-[3-(TRIFLUOROMETHYL)BENZYL]-1H-INDOLE-2-CARBOXAMIDE, Peroxisome proliferator-activated receptor gamma, SRC Peptide Fragment | | Authors: | Pokross, M.E, Evdokimov, A.G, Walter, R.L, Mekel, M.J, Hopkins, C.R. | | Deposit date: | 2006-06-25 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and synthesis of novel N-sulfonyl-2-indole carboxamides as potent PPAR-gamma binding agents with potential application to the treatment of osteoporosis.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2HFQ
 
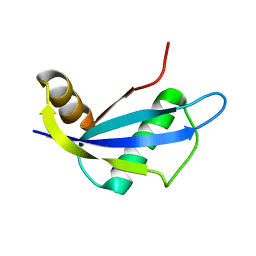 | | NMR structure of protein NE1680 from Nitrosomonas europaea: Northeast Structural Genomics Consortium target NeT5 | | Descriptor: | Hypothetical protein | | Authors: | Cort, J.R, Yee, A.A, Yim, V, Lukin, J, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-25 | | Release date: | 2006-09-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of protein NE1680 from Nitrosomonas europaea: Northeast Structural Genomics Consortium target NeT5
TO BE PUBLISHED
|
|
2HFR
 
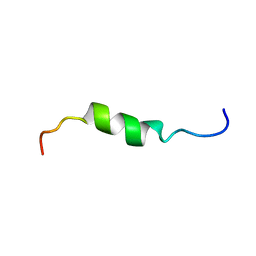 | | solution structure of antimicrobial peptide Fowlicidin 3 | | Descriptor: | Fowlicidin-3 | | Authors: | Bommineni, Y.R, Dai, H, Gong, Y, Prakash, O, Zhang, G. | | Deposit date: | 2006-06-26 | | Release date: | 2007-04-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Fowlicidin-3 is an alpha-helical cationic host defense peptide with potent antibacterial and lipopolysaccharide-neutralizing activities.
Febs J., 274, 2007
|
|
2HFS
 
 | |
2HFT
 
 | |
2HFU
 
 | |
2HFV
 
 | | Solution NMR Structure of Protein RPA1041 from Pseudomonas aeruginosa. Northeast Structural Genomics Consortium Target PaT90. | | Descriptor: | Hypothetical Protein RPA1041 | | Authors: | Eletsky, A, Atreya, H.S, Liu, G, Sukumaran, D, Garcia, M, Yee, A, Arrowsmith, C, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-26 | | Release date: | 2006-07-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Pseudomonas aeruginosa Hypothetical Protein RPA1041
TO BE PUBLISHED
|
|
2HFW
 
 | | Structural and kinetic analysis of proton shuttle residues in the active site of human carbonic anhydrase III | | Descriptor: | Carbonic anhydrase 3, ZINC ION | | Authors: | Elder, I, Fisher, S.Z, Laipis, P.J, Tu, C.K, McKenna, R, Silverman, D.N. | | Deposit date: | 2006-06-26 | | Release date: | 2007-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and kinetic analysis of proton shuttle residues in the active site of human carbonic anhydrase III.
Proteins, 68, 2007
|
|
2HFZ
 
 | |
2HG0
 
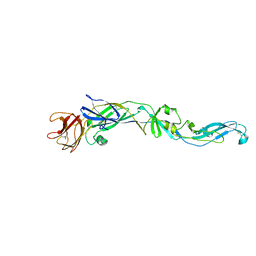 | | Structure of the West Nile Virus envelope glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein | | Authors: | Nybakken, G.E, Nelson, C.A, Chen, B.R, Diamond, M.S, Fremont, D.H. | | Deposit date: | 2006-06-26 | | Release date: | 2006-11-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the West Nile virus envelope glycoprotein.
J.Virol., 80, 2006
|
|
2HG1
 
 | |
2HG2
 
 | |
2HG3
 
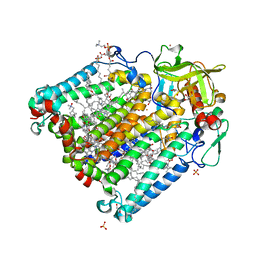 | | Reaction centre from Rhodobacter sphaeroides strain R-26.1 complexed with brominated phosphatidylcholine | | Descriptor: | (7R,14S)-14,15-DIBROMO-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2006-06-26 | | Release date: | 2007-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Brominated Lipids Identify Lipid Binding Sites on the Surface of the Reaction Center from Rhodobacter sphaeroides.
Biochemistry, 46, 2007
|
|
2HG4
 
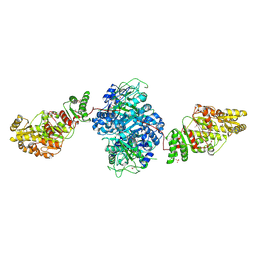 | | Structure of the ketosynthase-acyltransferase didomain of module 5 from DEBS. | | Descriptor: | 6-Deoxyerythronolide B Synthase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Tang, Y, Kim, C.Y, Mathews, I.I, Cane, D.E, Khosla, C. | | Deposit date: | 2006-06-26 | | Release date: | 2006-07-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The 2.7-A crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HG5
 
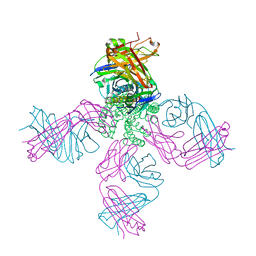 | | Cs+ complex of a K channel with an amide to ester substitution in the selectivity filter | | Descriptor: | (2S)-2-(BUTYRYLOXY)-3-HYDROXYPROPYL NONANOATE, CESIUM ION, FAB HEAVY CHAIN, ... | | Authors: | Valiyaveetil, F.I, MacKinnon, R, Muir, T.W. | | Deposit date: | 2006-06-26 | | Release date: | 2006-09-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural and Functional Consequences of an Amide-to-Ester Substitution in the Selectivity Filter of a Potassium Channel.
J.Am.Chem.Soc., 128, 2006
|
|
2HG6
 
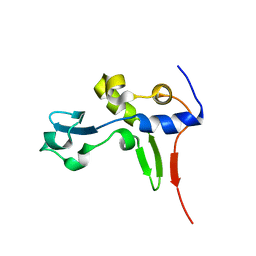 | | Solution NMR Structure of Protein PA1123 from Pseudomonas aeruginosa. Northeast Structural Genomics Consortium Target PaT4; Ontario Centre for Structural Proteomics Target PA1123. | | Descriptor: | Hypothetical protein | | Authors: | Lemak, A, Srisailam, S, Yee, A, Lukin, J.A, Orekhov, V.Y, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-26 | | Release date: | 2006-07-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a hypothetical protein from Pseudomonas aeruginosa (Northeast Structural Genomics Consortium Target: PaT4; Ontario Centre for Structural Proteomics Target: PA1123)
To be Published
|
|
2HG7
 
 | | Solution NMR structure of Phage-like element PBSX protein xkdW, Northeast Structural Genomics Consortium Target SR355 | | Descriptor: | Phage-like element PBSX protein xkdW | | Authors: | Liu, G, Parish, D, Xu, D, Atreya, H, Sukumaran, D, Ho, C.K, Jiang, M, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-26 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Phage-like element PBSX protein xkdW, Northeast Structural Genomics Consortium Target SR355
TO BE PUBLISHED
|
|
2HG8
 
 | |
2HG9
 
 | | Reaction centre from Rhodobacter sphaeroides strain R-26.1 complexed with tetrabrominated phosphatidylcholine | | Descriptor: | (7R,18S,19R)-18,19-DIBROMO-7-{[(9S,10S)-9,10-DIBROMOOCTADECANOYL]OXY}-4-HYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-P HOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2006-06-26 | | Release date: | 2007-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Brominated Lipids Identify Lipid Binding Sites on the Surface of the Reaction Center from Rhodobacter sphaeroides.
Biochemistry, 46, 2007
|
|
2HGA
 
 | | Solution NMR Structure of Conserved protein MTH1368, Northeast Structural Genomics Consortium Target TT821A | | Descriptor: | Conserved protein MTH1368 | | Authors: | Liu, G, Lin, Y, Parish, D, Shen, Y, Sukumaran, D, Yee, A, Semesi, A, Arrowsmith, C, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-26 | | Release date: | 2006-07-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Conserved protein MTH1368, Northeast Structural Genomics Consortium Target TT821A
TO BE PUBLISHED
|
|
2HGC
 
 | | Solution NMR structure of the YjcQ protein from Bacillus subtilis. Northeast Structural Genomics target SR346. | | Descriptor: | YjcQ protein | | Authors: | Rossi, P, Cort, J.R, Ho, C.K, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-26 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the YjcQ protein from Bacillus subtilis. Northeast Structural Genomics target SR346. (CASP Target)
To be Published
|
|
2HGD
 
 | |
2HGF
 
 | | HAIRPIN LOOP CONTAINING DOMAIN OF HEPATOCYTE GROWTH FACTOR, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HEPATOCYTE GROWTH FACTOR | | Authors: | Zhou, H, Mazzulla, M.J, Kaufman, J.D, Stahl, S.J, Wingfield, P.T, Rubin, J.S, Bottaro, D.P, Byrd, R.A. | | Deposit date: | 1997-12-18 | | Release date: | 1998-06-24 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of hepatocyte growth factor reveals a potential heparin-binding site.
Structure, 6, 1998
|
|
2HGH
 
 | |
2HGK
 
 | | Solution NMR Structure of protein YqcC from E. coli: Northeast Structural Genomics Consortium target ER225 | | Descriptor: | Hypothetical protein yqcC | | Authors: | Cort, J.R, Wang, D, Shetty, K, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-27 | | Release date: | 2007-06-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of protein YqcC from E.coli
To be Published
|
|
