2HBO
 
 | |
2HBP
 
 | | Solution Structure of Sla1 Homology Domain 1 | | Descriptor: | Cytoskeleton assembly control protein SLA1 | | Authors: | Overduin, M, Mahadev, R.K. | | Deposit date: | 2006-06-14 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of Sla1p homology domain 1 and interaction with the NPFxD endocytic internalization motif.
Embo J., 26, 2007
|
|
2HBQ
 
 | |
2HBR
 
 | |
2HBS
 
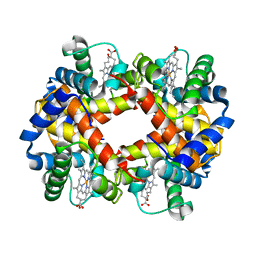 | | THE HIGH RESOLUTION CRYSTAL STRUCTURE OF DEOXYHEMOGLOBIN S | | Descriptor: | HEMOGLOBIN S (DEOXY), ALPHA CHAIN, BETA CHAIN, ... | | Authors: | Harrington, D.J, Adachi, K, Royer Junior, W.E. | | Deposit date: | 1997-05-06 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The high resolution crystal structure of deoxyhemoglobin S.
J.Mol.Biol., 272, 1997
|
|
2HBT
 
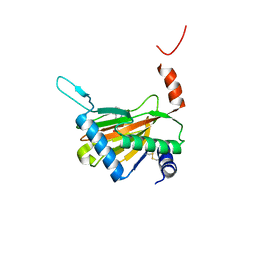 | | Crystal structure of HIF prolyl hydroxylase EGLN-1 in complex with a biologically active inhibitor | | Descriptor: | Egl nine homolog 1, FE (II) ION, N-[(1-CHLORO-4-HYDROXYISOQUINOLIN-3-YL)CARBONYL]GLYCINE | | Authors: | Evdokimov, A.G, Walter, R.L, Mekel, M, Pokross, M.E, Kawamoto, R, Boyer, A. | | Deposit date: | 2006-06-14 | | Release date: | 2006-06-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of HIF prolyl hydroxylase in complex with a biologically active inhibitor
To be Published
|
|
2HBU
 
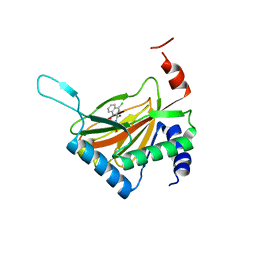 | | Crystal structure of HIF prolyl hydroxylase EGLN-1 in complex with a biologically active inhibitor | | Descriptor: | Egl nine homolog 1, FE (II) ION, N-[(1-CHLORO-4-HYDROXYISOQUINOLIN-3-YL)CARBONYL]GLYCINE | | Authors: | Evdokimov, A.G, Walter, R.L, Mekel, M, Pokross, M.E, Kawamoto, R, Boyer, A. | | Deposit date: | 2006-06-14 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of HIF prolyl hydroxylase in complex with a biologically active inhibitor
To be Published
|
|
2HBV
 
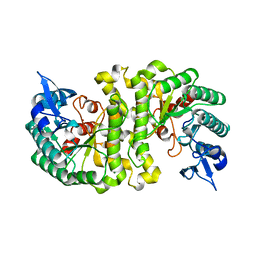 | | Crystal Structure of alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde-Decarboxylase (ACMSD) | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, MAGNESIUM ION, ZINC ION | | Authors: | Martynowski, D, Eyobo, Y, Li, T, Yang, K, Liu, A, Zhang, H. | | Deposit date: | 2006-06-14 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of alpha-Amino-beta-carboxymuconate-epsilon-semialdehyde Decarboxylase: Insight into the Active Site and Catalytic Mechanism of a Novel Decarboxylation Reaction.
Biochemistry, 45, 2006
|
|
2HBW
 
 | |
2HBX
 
 | | Crystal Structure of alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde-Decarboxylase (ACMSD) | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, COBALT (II) ION | | Authors: | Martynowski, D, Eyobo, Y, Li, T, Yang, K, Liu, A, Zhang, H. | | Deposit date: | 2006-06-14 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of alpha-Amino-beta-carboxymuconate-epsilon-semialdehyde Decarboxylase: Insight into the Active Site and Catalytic Mechanism of a Novel Decarboxylation Reaction.
Biochemistry, 45, 2006
|
|
2HBY
 
 | |
2HBZ
 
 | |
2HC0
 
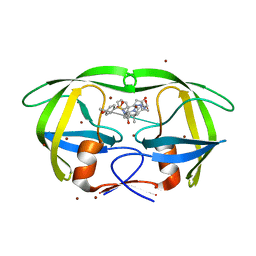 | | Structure of HIV protease 6X mutant in complex with AB-2. | | Descriptor: | BROMIDE ION, Protease, [1-((1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL)-1H-1,2,3-TRIAZOL-4-YL]METHYL (1R,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YLCARBAMATE | | Authors: | Heaslet, H, Brik, A, Lin, Y.-C, Elder, J.H, Stout, C.D. | | Deposit date: | 2006-06-14 | | Release date: | 2007-06-26 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of HIV Protease 6X Mutant in complex with AB-2
To be Published
|
|
2HC1
 
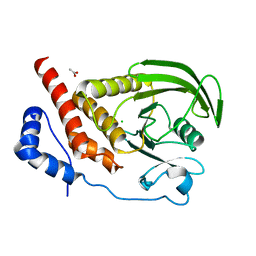 | | Engineered catalytic domain of protein tyrosine phosphatase HPTPbeta. | | Descriptor: | ACETATE ION, CHLORIDE ION, Receptor-type tyrosine-protein phosphatase beta | | Authors: | Evdokimov, A.G, Pokross, M, Walter, R, Mekel, M. | | Deposit date: | 2006-06-14 | | Release date: | 2006-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Engineering the catalytic domain of human protein tyrosine phosphatase beta for structure-based drug discovery.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2HC2
 
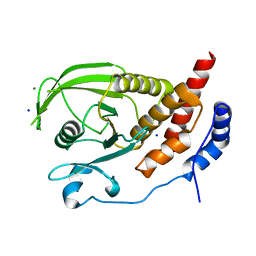 | | Engineered protein tyrosine phosphatase beta catalytic domain | | Descriptor: | MAGNESIUM ION, Receptor-type tyrosine-protein phosphatase beta, SODIUM ION | | Authors: | Evdokimov, A.G, Pokross, M, Walter, R, Mekel, M. | | Deposit date: | 2006-06-15 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineering the catalytic domain of human protein tyrosine phosphatase beta for structure-based drug discovery.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2HC4
 
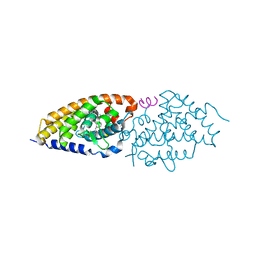 | | Crystal structure of the LBD of VDR of Danio rerio in complex with calcitriol | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, SRC-1 from Nuclear receptor coactivator 1, Vitamin D receptor | | Authors: | Ciesielski, F, Rochel, N, Moras, D. | | Deposit date: | 2006-06-15 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Adaptability of the Vitamin D nuclear receptor to the synthetic ligand Gemini: remodelling the LBP with one side chain rotation
J.Steroid Biochem.Mol.Biol., 103, 2007
|
|
2HC5
 
 | | Solution NMR Structure of Protein yvyC from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR482. | | Descriptor: | Hypothetical protein yvyC | | Authors: | Eletsky, A, Liu, G, Atreya, H.S, Sukumaran, D.K, Wang, D, Cunningham, K, Janjua, H, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-15 | | Release date: | 2006-08-15 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of protein YvyC from Bacillus subtilis reveals unexpected structural similarity between two PFAM families.
Proteins, 76, 2009
|
|
2HC7
 
 | | 2'-selenium-T A-DNA [G(TSe)GTACAC] | | Descriptor: | 5'-D(*GP*(2ST)P*GP*TP*AP*CP*AP*C)-3' | | Authors: | Jiang, J, Sheng, J, Huang, Z. | | Deposit date: | 2006-06-15 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of 2'-Selenium modified T A-DNA G(TSe)GTACAC
TO BE PUBLISHED
|
|
2HC8
 
 | | Structure of the A. fulgidus CopA A-domain | | Descriptor: | Cation-transporting ATPase, P-type | | Authors: | Sazinsky, M.H, Agawal, S, Arguello, J.M, Rosenzweig, A.C. | | Deposit date: | 2006-06-15 | | Release date: | 2006-09-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the Actuator Domain from the Archaeoglobus fulgidus Cu(+)-ATPase(,).
Biochemistry, 45, 2006
|
|
2HC9
 
 | | Structure of Caenorhabditis elegans leucine aminopeptidase-zinc complex (LAP1) | | Descriptor: | BICARBONATE ION, GLYCEROL, Leucine aminopeptidase 1, ... | | Authors: | Zhan, C, Patskovsky, Y, Wengerter, B.C, Ramagopal, U, Milstein, S, Vidal, M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-06-15 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure and Function of Caenorhabditis Elegans Leucine Aminopeptidase
To be Published
|
|
2HCA
 
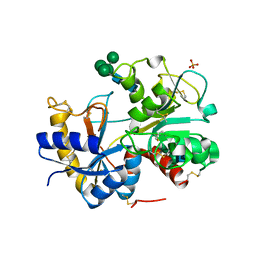 | | Crystal structure of bovine lactoferrin C-lobe liganded with Glucose at 2.8 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Mir, R, Prem Kumar, R, Ethayathulla, A.S, Singh, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2006-06-15 | | Release date: | 2006-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of bovine lactoferrin C-lobe liganded with Glucose at 2.8 A resolution
To be Published
|
|
2HCB
 
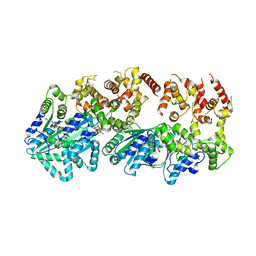 | | Structure of AMPPCP-bound DnaA from Aquifex aeolicus | | Descriptor: | Chromosomal replication initiator protein dnaA, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Erzberger, J.P, Mott, M.L, Berger, J.M. | | Deposit date: | 2006-06-15 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2HCC
 
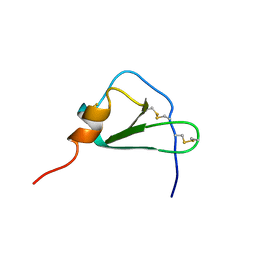 | | SOLUTION STRUCTURE OF THE HUMAN CHEMOKINE HCC-2, NMR, 30 STRUCTURES | | Descriptor: | HUMAN CHEMOKINE HCC-2 | | Authors: | Sticht, H, Escher, S.E, Schweimer, K, Forssmann, W.G, Roesch, P, Adermann, K. | | Deposit date: | 1998-07-03 | | Release date: | 1999-07-13 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the human CC chemokine 2: A monomeric representative of the CC chemokine subtype.
Biochemistry, 38, 1999
|
|
2HCD
 
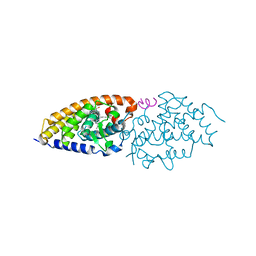 | | Crystal structure of the ligand binding domain of the Vitamin D nuclear receptor in complex with Gemini and a coactivator peptide | | Descriptor: | 21-NOR-9,10-SECOCHOLESTA-5,7,10(19)-TRIENE-1,3,25-TRIOL, 20-(4-HYDROXY-4-METHYLPENTYL)-, (1A,3B,5Z,7E), ... | | Authors: | Ciesielski, F, Rochel, N, Moras, D. | | Deposit date: | 2006-06-16 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Adaptability of the Vitamin D nuclear receptor to the synthetic ligand Gemini: remodelling the LBP with one side chain rotation
J.Steroid Biochem.Mol.Biol., 103, 2007
|
|
2HCF
 
 | |
