7PCF
 
 | | Human methemoglobin bound to Staphylococcus aureus hemophore IsdB | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, Iron-regulated surface determinant protein B, ... | | Authors: | De Bei, O, Gianquinto, E, Chirgadze, D.Y, Hardwick, S.W, Spyrakis, F, Luisi, B.F, Campanini, B. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (5.82 Å) | | Cite: | Cryo-EM structures of staphylococcal IsdB bound to human hemoglobin reveal the process of heme extraction.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7PCQ
 
 | | Human carboxyhemoglobin bound to Staphylococcus aureus hemophore IsdB - 1:1 complex | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, Iron-regulated surface determinant protein B, ... | | Authors: | De Bei, O, Gianquinto, E, Chirgadze, D.Y, Hardwick, S.W, Spyrakis, F, Luisi, B.F, Campanini, B. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Cryo-EM structures of staphylococcal IsdB bound to human hemoglobin reveal the process of heme extraction.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
830C
 
 | | COLLAGENASE-3 (MMP-13) COMPLEXED TO A SULPHONE-BASED HYDROXAMIC ACID | | Descriptor: | 4-[4-(4-CHLORO-PHENOXY)-BENZENESULFONYLMETHYL]-TETRAHYDRO-PYRAN-4-CARBOXYLIC ACID HYDROXYAMIDE, CALCIUM ION, MMP-13, ... | | Authors: | Lovejoy, B, Welch, A, Carr, S, Luong, C, Broka, C, Hendricks, R.T, Campbell, J, Walker, K, Martin, R, Van Wart, H, Browner, M.F. | | Deposit date: | 1998-08-06 | | Release date: | 1999-08-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of MMP-1 and -13 reveal the structural basis for selectivity of collagenase inhibitors.
Nat.Struct.Biol., 6, 1999
|
|
7PHR
 
 | | Structure of a fully assembled T-cell receptor engaging a tumor-associated peptide-MHC I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Susac, L, Thomas, C, Tampe, R. | | Deposit date: | 2021-08-18 | | Release date: | 2022-08-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structure of a fully assembled tumor-specific T cell receptor ligated by pMHC.
Cell, 185, 2022
|
|
7P63
 
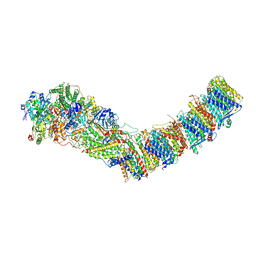 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 6, Closed state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7P69
 
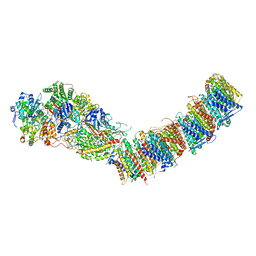 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 6, Resting state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7P7K
 
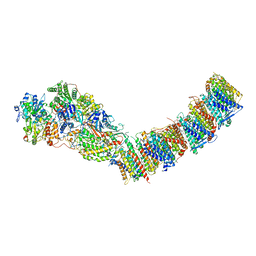 | | Complex I from E. coli, DDM/LMNG-purified, with DQ, Resting state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, EICOSANE, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7P7L
 
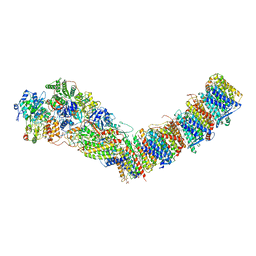 | | Complex I from E. coli, DDM/LMNG-purified, with NADH and FMN, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, EICOSANE, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7P62
 
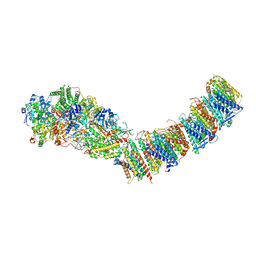 | | Complex I from E. coli, DDM-purified, Apo, Resting state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7P61
 
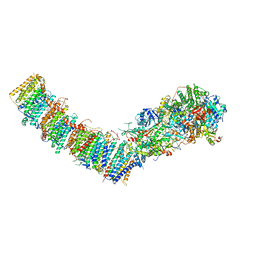 | | Complex I from E. coli, DDM-purified, with NADH, Resting state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7P64
 
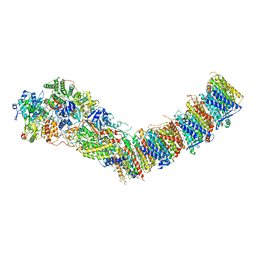 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 6, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7P7C
 
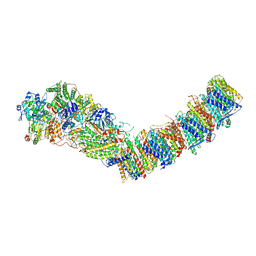 | | Complex I from E. coli, DDM/LMNG-purified, Apo, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, EICOSANE, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7P7E
 
 | | Complex I from E. coli, DDM/LMNG-purified, Apo, Resting state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, EICOSANE, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7P7J
 
 | | Complex I from E. coli, DDM/LMNG-purified, with DQ, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7P7M
 
 | | Complex I from E. coli, DDM/LMNG-purified, inhibited by Piericidin A, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, EICOSANE, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7PBU
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvA-HJ core [t2 dataset] | | Descriptor: | Holliday junction, Holliday junction ATP-dependent DNA helicase RuvA | | Authors: | Goessweiner-Mohr, N, Fahrenkamp, D, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
6EZ6
 
 | | PI3 kinase delta in complex with Methyl 5-(4-(5-((4-isopropylpiperazin-1-yl)methyl)oxazol-2-yl)-1H-indazol-6-yl)-2-methoxynicotinate | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, methyl 2-methoxy-5-[4-[5-[(4-propan-2-ylpiperazin-1-yl)methyl]-1,3-oxazol-2-yl]-2~{H}-indazol-6-yl]pyridine-3-carboxylate | | Authors: | Convery, M.A, Campos, S, Dalton, S.E. | | Deposit date: | 2017-11-14 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Selectively Targeting the Kinome-Conserved Lysine of PI3K delta as a General Approach to Covalent Kinase Inhibition.
J. Am. Chem. Soc., 140, 2018
|
|
7PBT
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s1 [t1 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Wald, J, Fahrenkamp, D, Goessweiner-Mohr, N, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
7PBQ
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s0+A [t2 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Goessweiner-Mohr, N, Fahrenkamp, D, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
7PBS
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s0+A [t1 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Goessweiner-Mohr, N, Fahrenkamp, D, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
7PBR
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s0-A [t2 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvB, MAGNESIUM ION, ... | | Authors: | Goessweiner-Mohr, N, Fahrenkamp, D, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
8ATJ
 
 | | Crystal Structure of Shank2-SAM domain | | Descriptor: | CHLORIDE ION, FORMIC ACID, Isoform 4 of SH3 and multiple ankyrin repeat domains protein 2, ... | | Authors: | Bento, I, Gracia Alai, M, Kreienkamp, J.-H. | | Deposit date: | 2022-08-23 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.117 Å) | | Cite: | Structural deficits in key domains of Shank2 lead to alterations in postsynaptic nanoclusters and to a neurodevelopmental disorder in humans.
Mol Psychiatry, 2022
|
|
8B10
 
 | | Crystal Structure of Shank2-SAM mutant domain - L1800W | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bento, I, Gracia Alai, M, Kreienkamp, J.-H. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-30 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural deficits in key domains of Shank2 lead to alterations in postsynaptic nanoclusters and to a neurodevelopmental disorder in humans.
Mol Psychiatry, 29, 2024
|
|
3EVP
 
 | | crystal structure of circular-permutated EGFP | | Descriptor: | Green fluorescent protein,Green fluorescent protein | | Authors: | Wang, Q, Shui, B, Kotlikoff, M.I, Sondermann, H. | | Deposit date: | 2008-10-13 | | Release date: | 2008-12-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.453 Å) | | Cite: | Structural Basis for Calcium Sensing by GCaMP2.
Structure, 16, 2008
|
|
6Y86
 
 | |
