1EYX
 
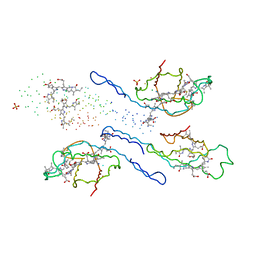 | | CRYSTAL STRUCTURE OF R-PHYCOERYTHRIN AT 2.2 ANGSTROMS | | Descriptor: | PHYCOCYANOBILIN, PHYCOUROBILIN, R-PHYCOERYTHRIN, ... | | Authors: | Contreras-Martel, C, Legrand, P, Piras, C, Vernede, X, Martinez-Oyanedel, J, Bunster, M, Fontecilla-Camps, J.C. | | Deposit date: | 2000-05-09 | | Release date: | 2000-11-22 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallization and 2.2 A resolution structure of R-phycoerythrin from Gracilaria chilensis: a case of perfect hemihedral twinning.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
6CAE
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with NOSO-95179 antibiotic and bound to mRNA and A-, P- and E-site tRNAs at 2.6A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Pantel, L, Florin, T, Dobosz-Bartoszek, M, Racine, E, Sarciaux, M, Serri, M, Houard, J, Campagne, J.M, Marcia de Figueiredo, R, Midrier, C, Gaudriault, S, Givaudan, A, Lanois, A, Forst, S, Aumelas, A, Cotteaux-Lautard, C, Bolla, J.M, Vingsbo Lundberg, C, Huseby, D, Hughes, D, Villain-Guillot, P, Mankin, A.S, Polikanov, Y.S, Gualtieri, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-04-18 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Odilorhabdins, Antibacterial Agents that Cause Miscoding by Binding at a New Ribosomal Site.
Mol. Cell, 70, 2018
|
|
6VAF
 
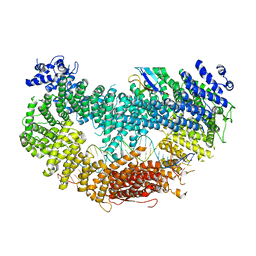 | |
6VAE
 
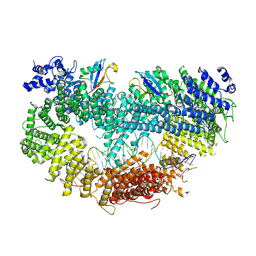 | |
6V79
 
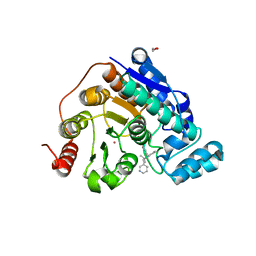 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 (CD2) complexed with NF2376 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(2S)-3,3-dimethyl-2-(pyridin-3-yl)-2,3-dihydro-1H-indol-1-yl]methyl}-N-hydroxybenzamide, Hdac6 protein, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2019-12-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.03951526 Å) | | Cite: | Harnessing the Role of HDAC6 in Idiopathic Pulmonary Fibrosis: Design, Synthesis, Structural Analysis, and Biological Evaluation of Potent Inhibitors.
J.Med.Chem., 64, 2021
|
|
6UYH
 
 | |
7QIA
 
 | | Structure of apo-EleNRMT in complex with two nanobodies at 3.5A | | Descriptor: | Divalent metal cation transporter, Nanobody 1, Nanobody 2 | | Authors: | Ramanadane, K, Straub, M.S, Dutzler, R, Manatschal, C. | | Deposit date: | 2021-12-14 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and functional properties of a magnesium transporter of the SLC11/NRAMP family.
Elife, 11, 2022
|
|
7QIC
 
 | | Structure of magnesium-bound EleNRMT in complex with two nanobodies at 4.1A | | Descriptor: | Divalent metal cation transporter, MAGNESIUM ION, Nanobody 1, ... | | Authors: | Ramanadane, K, Straub, M.S, Dutzler, R, Manatschal, C. | | Deposit date: | 2021-12-14 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural and functional properties of a magnesium transporter of the SLC11/NRAMP family.
Elife, 11, 2022
|
|
6YST
 
 | | Structure of the P+9 ArfB-ribosome complex with P/E hybrid tRNA in the post-hydrolysis state | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Chan, K.-H, Petrychenko, V, Mueller, C, Maracci, C, Holtkamp, W, Wilson, D.N, Fischer, N, Rodnina, M.V. | | Deposit date: | 2020-04-23 | | Release date: | 2020-08-19 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of ribosome rescue by alternative ribosome-rescue factor B.
Nat Commun, 11, 2020
|
|
6YSR
 
 | | Structure of the P+9 stalled ribosome complex | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Chan, K.-H, Petrychenko, V, Mueller, C, Maracci, C, Holtkamp, W, Wilson, D.N, Fischer, N, Rodnina, M.V. | | Deposit date: | 2020-04-23 | | Release date: | 2020-08-19 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of ribosome rescue by alternative ribosome-rescue factor B.
Nat Commun, 11, 2020
|
|
6YSU
 
 | | Structure of the P+0 ArfB-ribosome complex in the post-hydrolysis state | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Chan, K.-H, Petrychenko, V, Mueller, C, Maracci, C, Holtkamp, W, Wilson, D.N, Fischer, N, Rodnina, M.V. | | Deposit date: | 2020-04-23 | | Release date: | 2020-08-19 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanism of ribosome rescue by alternative ribosome-rescue factor B.
Nat Commun, 11, 2020
|
|
7UO7
 
 | | SARS-CoV-2 replication-transcription complex bound to ATP, in a pre-catalytic state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
7UOE
 
 | | SARS-CoV-2 replication-transcription complex bound to CTP, in a pre-catalytic state | | Descriptor: | 3'-DEOXYURIDINE-5'-MONOPHOSPHATE, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
7UO4
 
 | | SARS-CoV-2 replication-transcription complex bound to Remdesivir triphosphate, in a pre-catalytic state | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
7UO9
 
 | | SARS-CoV-2 replication-transcription complex bound to UTP, in a pre-catalytic state | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
7UOB
 
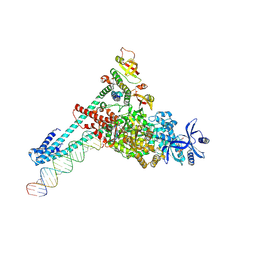 | | SARS-CoV-2 replication-transcription complex bound to GTP, in a pre-catalytic state | | Descriptor: | 3'-DEOXYURIDINE-5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
7RDX
 
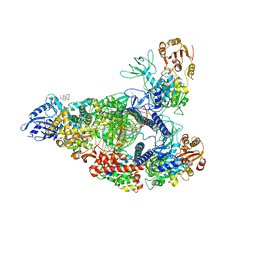 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - open class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-24 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE1
 
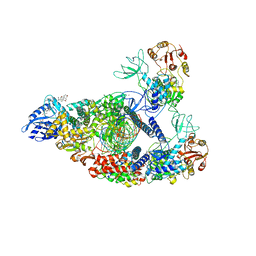 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC (composite) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RDZ
 
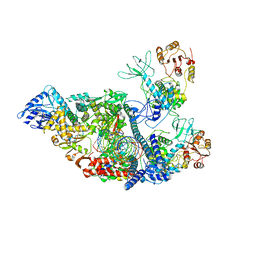 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - apo class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Helicase, MAGNESIUM ION, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-24 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE3
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC dimer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-24 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE2
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(1)-RTC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RDY
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - engaged class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE0
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - swiveled class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Helicase, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7R5O
 
 | |
6YSS
 
 | | Structure of the P+9 ArfB-ribosome complex in the post-hydrolysis state | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Chan, K.-H, Petrychenko, V, Mueller, C, Maracci, C, Holtkamp, W, Wilson, D.N, Fischer, N, Rodnina, M.V. | | Deposit date: | 2020-04-23 | | Release date: | 2020-08-19 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Mechanism of ribosome rescue by alternative ribosome-rescue factor B.
Nat Commun, 11, 2020
|
|
