9HPH
 
 | | Protein kinase CK2 bound to KDX1381 | | Descriptor: | CHLORIDE ION, Casein kinase II subunit alpha, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Krimm, I. | | Deposit date: | 2024-12-13 | | Release date: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Discovery of KDX1381, a Bivalent CK2 alpha Inhibitor for the Treatment of Solid Tumors as a Single Agent or in Combination.
J.Med.Chem., 68, 2025
|
|
5W63
 
 | | Crystal structure of channel catfish BAX | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator bax, SULFATE ION | | Authors: | Robin, A.Y, Colman, P.M, Czabotar, P.E, Luo, C.S. | | Deposit date: | 2017-06-16 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.436 Å) | | Cite: | Ensemble Properties of Bax Determine Its Function.
Structure, 26, 2018
|
|
6DLK
 
 | |
6S6G
 
 | |
6ITJ
 
 | | Crystal structure of FGFR1 kinase domain in complex with compound 3 | | Descriptor: | 4-azanyl-3-(3,5-dimethyl-1-benzofuran-2-yl)-2-phenyl-6~{H}-pyrazolo[3,4-d]pyridazin-7-one, Fibroblast growth factor receptor 1 | | Authors: | Xu, Y, Liu, Q. | | Deposit date: | 2018-11-23 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
6T0G
 
 | | Crystal structure of CYP124 in complex with vitamin D3 | | Descriptor: | (1S,3Z)-3-[(2E)-2-[(1R,3AR,7AS)-7A-METHYL-1-[(2R)-6-METHYLHEPTAN-2-YL]-2,3,3A,5,6,7-HEXAHYDRO-1H-INDEN-4-YLIDENE]ETHYLI DENE]-4-METHYLIDENE-CYCLOHEXAN-1-OL, MAGNESIUM ION, Methyl-branched lipid omega-hydroxylase, ... | | Authors: | Bukhdruker, S, Marin, E, Varaksa, T, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Metabolic Fate of Human Immunoactive Sterols in Mycobacterium tuberculosis.
J.Mol.Biol., 433, 2021
|
|
7ZQ9
 
 | | Dimeric PSI of Chlamydomonas reinhardtii at 2.74 A resolution (symmetry expanded) | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Naschberger, A, Amunts, A. | | Deposit date: | 2022-04-29 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Algal photosystem I dimer and high-resolution model of PSI-plastocyanin complex.
Nat.Plants, 8, 2022
|
|
1NAA
 
 | | Cellobiose Dehydrogenase Flavoprotein Fragment in Complex with Cellobionolactam | | Descriptor: | (2R,3R,4R,5R)-4,5-dihydroxy-2-(hydroxymethyl)-6-oxopiperidin-3-yl beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-HYDROXY-FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hallberg, B.M, Henriksson, G, Pettersson, G, Vasella, A, Divne, C. | | Deposit date: | 2002-11-27 | | Release date: | 2003-01-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of the reductive half-reaction in cellobiose dehydrogenase
J.BIOL.CHEM., 278, 2003
|
|
7BZG
 
 | | Structure of Bacillus subtilis HxlR, wild type in complex with formaldehyde and DNA | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*CP*AP*GP*TP*AP*TP*CP*CP*TP*CP*GP*AP*GP*GP*AP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Zhu, R, Chen, P.R. | | Deposit date: | 2020-04-27 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Genetically encoded formaldehyde sensors inspired by a protein intra-helical crosslinking reaction.
Nat Commun, 12, 2021
|
|
7ZQD
 
 | | Dimeric PSI of Chlamydomonas reinhardtii at 2.97 A resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Naschberger, A, Amunts, A. | | Deposit date: | 2022-04-29 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Algal photosystem I dimer and high-resolution model of PSI-plastocyanin complex.
Nat.Plants, 8, 2022
|
|
7ZQC
 
 | | Monomeric PSI of Chlamydomonas reinhardtii at 2.31 A resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Naschberger, A, Amunts, A. | | Deposit date: | 2022-04-29 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | Algal photosystem I dimer and high-resolution model of PSI-plastocyanin complex.
Nat.Plants, 8, 2022
|
|
5IU4
 
 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with ZM241385 at 1.7A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Segala, E, Guo, D, Cheng, R.K.Y, Bortolato, A, Deflorian, F, Dore, A.S, Errey, J.C, Heitman, L.H, Ijzerman, A.P, Marshall, F.H, Cooke, R.M. | | Deposit date: | 2016-03-17 | | Release date: | 2016-06-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Controlling the Dissociation of Ligands from the Adenosine A2A Receptor through Modulation of Salt Bridge Strength.
J.Med.Chem., 59, 2016
|
|
9FMI
 
 | | PsiM N247A in complex with SAH and norbaeocystin | | Descriptor: | CHLORIDE ION, Norbaeocystin, Psilocybin synthase, ... | | Authors: | Hudspeth, J, Rupp, B, Werten, S. | | Deposit date: | 2024-06-06 | | Release date: | 2024-10-23 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | The Second Methylation in Psilocybin Biosynthesis Is Enabled by a Hydrogen Bonding Network Extending into the Secondary Sphere Surrounding the Methyltransferase Active Site.
Chembiochem, 25, 2024
|
|
5FFF
 
 | |
6IER
 
 | | Apo structure of a beta-glucosidase 1317 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, beta-glucosidase 1317 | | Authors: | Xie, W, Liu, X. | | Deposit date: | 2018-09-16 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Improving the cellobiose-hydrolysis activity and glucose-tolerance of a thermostable beta-glucosidase through rational design.
Int.J.Biol.Macromol., 136, 2019
|
|
6G6Z
 
 | | Eg5-inhibitor complex | | Descriptor: | (~{N}~{Z})-~{N}-[(5~{S})-4-ethanoyl-5-methyl-5-phenyl-1,3,4-thiadiazolidin-2-ylidene]ethanamide, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Talapatra, S.K, Kozielski, F, Tham, C.L. | | Deposit date: | 2018-04-04 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Eg5 - K858 complex and implications for structure-based design of thiadiazole-containing inhibitors.
Eur J Med Chem, 156, 2018
|
|
5VZE
 
 | | Post-catalytic complex of human Polymerase Mu (W434A) mutant with incoming UTP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*GP*GP*CP*AP*TP*AP*CP*G)-3'), ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2017-05-27 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.506 Å) | | Cite: | Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu.
Nucleic Acids Res., 45, 2017
|
|
6QTN
 
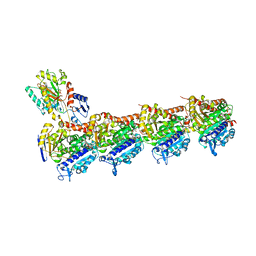 | | Tubulin-cyclostreptin complex | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Balaguer, F.d.A, Muehlethaler, T, Estevez-Gallego, J, Calvo, E, Gimenez-Abian, J.F, Risinger, A.L, Sorensen, E.J, Vanderwal, C.D, Altmann, K.-H, Mooberry, S.L, Steinmetz, M.O, Oliva, M.A, Prota, A.E, Diaz, J.F. | | Deposit date: | 2019-02-25 | | Release date: | 2019-04-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Cyclostreptin-Tubulin Adduct: Implications for Tubulin Activation by Taxane-Site Ligands.
Int J Mol Sci, 20, 2019
|
|
6ZXS
 
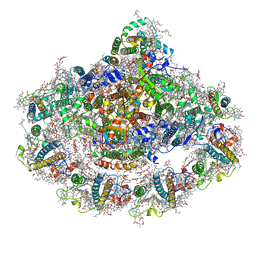 | | Cold grown Pea Photosystem I | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Caspy, I, Borovikova-Sheinker, A, Subramanyam, R, Nelson, N. | | Deposit date: | 2020-07-30 | | Release date: | 2021-07-07 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of cold grown pea Photosystem I
To Be Published
|
|
7SV2
 
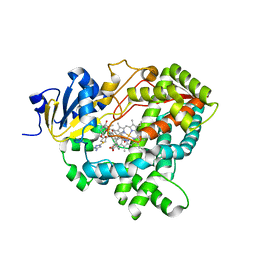 | | Human Cytochrome P450 (CYP) 3A5 ternary complex with azamulin | | Descriptor: | (3aS,4R,5S,6R,8R,9R,9aR,10R)-6-ethyl-5-hydroxy-4,6,9,10-tetramethyl-1-oxodecahydro-3a,9-propanocyclopenta[8]annulen-8-yl [(5-amino-1H-1,2,4-triazol-3-yl)sulfanyl]acetate, Cytochrome P450 3A5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hsu, M, Johnson, E.F. | | Deposit date: | 2021-11-18 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural characterization of the homotropic cooperative binding of azamulin to human cytochrome P450 3A5.
J.Biol.Chem., 298, 2022
|
|
5CPF
 
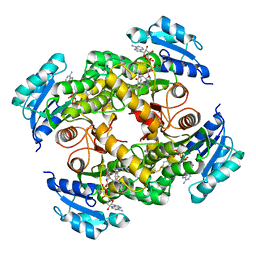 | | Compensation of the effect of isoleucine to alanine mutation by designed inhibition in the InhA enzyme | | Descriptor: | 2-(2-methylphenoxy)-5-[(4-phenyl-1H-1,2,3-triazol-1-yl)methyl]phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.-J, Lai, C.-T, Pan, P, Yu, W, Shah, S, Bommineni, G.R, Perrone, V, Garcia-Diaz, M, Tonge, P.J, Simmerling, C. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.409 Å) | | Cite: | Rational Modulation of the Induced-Fit Conformational Change for Slow-Onset Inhibition in Mycobacterium tuberculosis InhA.
Biochemistry, 54, 2015
|
|
6C9P
 
 | |
4S22
 
 | | Crystal structure of K29 linked di-Ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, IODIDE ION, ... | | Authors: | Kristariyanto, Y.A, Abdul Rehman, S.A, Campbell, D.G, Morrice, N.A, Johnson, C, Toth, R, Kulathu, Y. | | Deposit date: | 2015-01-17 | | Release date: | 2015-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | K29-selective ubiquitin binding domain reveals structural basis of specificity and heterotypic nature of k29 polyubiquitin.
Mol.Cell, 58, 2015
|
|
9H8Q
 
 | | Eugenol Oxidase (EUGO) from Rhodococcus jostii RHA1, mutant DTT-T425G | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Rozeboom, H.J, Fraaije, M.W. | | Deposit date: | 2024-10-29 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biocatalytic Cleavage of para-Acetoxy Benzyl Ethers: Application to Protecting Group Chemistry
Acs Catalysis, 15, 2025
|
|
9ILF
 
 | |
