4RGT
 
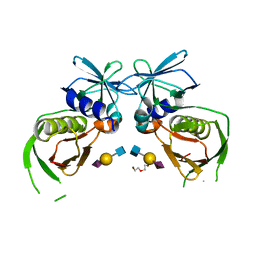 | | 2.0 Angstrom Crystal Structure of Superantigen-like Protein from Staphylococcus aureus in Complex with 3-N-Acetylneuraminyl-N-acetyllactosamine. | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Putative uncharacterized protein | | Authors: | Minasov, G, Nocadello, S, Shuvalova, L, Filippova, E.V, Halavaty, A, Dubrovska, I, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-30 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 Angstrom Crystal Structure of Superantigen-like Protein from Staphylococcus aureus in Complex with 3-N-Acetylneuraminyl-N-acetyllactosamine.
TO BE PUBLISHED
|
|
4RH6
 
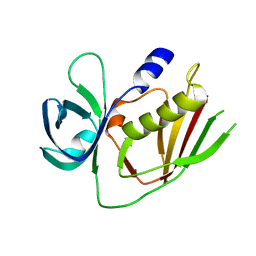 | | 2.9 Angstrom Crystal Structure of Putative Exotoxin 3 from Staphylococcus aureus. | | Descriptor: | CHLORIDE ION, Exotoxin 3, putative | | Authors: | Minasov, G, Nocadello, S, Shuvalova, L, Filippova, E.V, Halavaty, A, Dubrovska, I, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-01 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 2.9 Angstrom Crystal Structure of Putative Exotoxin 3 from Staphylococcus aureus.
TO BE PUBLISHED
|
|
4RCO
 
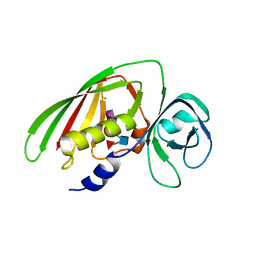 | | 1.9 Angstrom Crystal Structure of Superantigen-like Protein, Exotoxin from Staphylococcus aureus, in Complex with Sialyl-LewisX. | | Descriptor: | CHLORIDE ION, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose, Putative uncharacterized protein | | Authors: | Minasov, G, Nocadello, S, Shuvalova, L, Filippova, E, Halavaty, A, Dubrovska, I, Flores, K, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-16 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Crystal Structure of Superantigen-like Protein, Exotoxin from Staphylococcus aureus, in Complex with Sialyl-LewisX.
TO BE PUBLISHED
|
|
6GX4
 
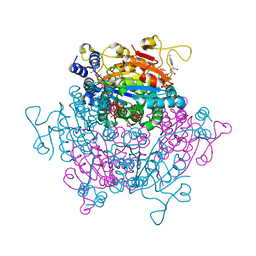 | | The molybdenum storage protein: with ATP/Mn2+ and with POM clusters formed under in vitro conditions | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, MOLYBDATE ION, ... | | Authors: | Poppe, J, Bruenle, S, Hail, R, Ermler, E. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Molybdenum Storage Protein: A soluble ATP hydrolysis-dependent molybdate pump.
FEBS J., 285, 2018
|
|
8BGR
 
 | |
7ML0
 
 | | RNA polymerase II pre-initiation complex (PIC1) | | Descriptor: | BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7ML1
 
 | | RNA polymerase II pre-initiation complex (PIC2) | | Descriptor: | BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7ML4
 
 | | RNA polymerase II initially transcribing complex (ITC) | | Descriptor: | BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7ML3
 
 | | General transcription factor TFIIH (weak binding) | | Descriptor: | BJ4_G0050160.mRNA.1.CDS.1, General transcription and DNA repair factor IIH, General transcription and DNA repair factor IIH helicase subunit XPB, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7ML2
 
 | | RNA polymerase II pre-initiation complex (PIC3) | | Descriptor: | BJ4_G0004860.mRNA.1.CDS.1, BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7N9I
 
 | | 1.4A Structure of Drosophila melanogaster Frataxin | | Descriptor: | Frataxin homolog, mitochondrial, SULFATE ION | | Authors: | Brunzelle, J.S, Stemmler, T.L. | | Deposit date: | 2021-06-17 | | Release date: | 2022-06-22 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Drosophila melanogaster frataxin: protein crystal and predicted solution structure with identification of the iron-binding regions.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6Y4D
 
 | | Crystal structure of a short-chain dehydrogenase/reductase (SDR) from Zephyranthes treatiae in complex with NADP+ | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, short-chain dehydrogenase/reductase (SDR) | | Authors: | Sautner, V, Steimle, S, Roth, S, Mueller, M, Tittmann, K. | | Deposit date: | 2020-02-20 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crossing the Border: From Keto- to Imine Reduction in Short-Chain Dehydrogenases/Reductases.
Chembiochem, 21, 2020
|
|
6GGU
 
 | | Crystal structure of native FE-hydrogenase from Methanothermobacter marburgensis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5,10-methenyltetrahydromethanopterin hydrogenase, GLYCEROL, ... | | Authors: | Wagner, T, Huang, G, Ermler, U, Shima, S. | | Deposit date: | 2018-05-03 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Self-protection of the catalytic iron center of a methanogenic [Fe]-hydrogenase via a dynamic dimer-to-hexamer transformation
To Be Published
|
|
6HUZ
 
 | | HmdII from Desulfurobacterium thermolithotrophum reconstituted with Fe-guanylylpyridinol (FeGP) cofactor and co-crystallized with methenyl-tetrahydrofolate form B | | Descriptor: | 1,2-ETHANEDIOL, 5,10-Methenyltetrahydrofolate, Coenzyme F420-dependent N(5),N(10)-methenyltetrahydromethanopterin reductase-related protein, ... | | Authors: | Watanabe, T, Wagner, T, Huang, G, Kahnt, J, Ataka, K, Ermler, U, Shima, S. | | Deposit date: | 2018-10-09 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Bacterial [Fe]-Hydrogenase Paralog HmdII Uses Tetrahydrofolate Derivatives as Substrates.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
6HAC
 
 | | Crystal structure of [Fe]-hydrogenase (Hmd) holoenzyme from Methanococcus aeolicus (open form) | | Descriptor: | 5,10-methenyltetrahydromethanopterin hydrogenase, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Huang, G, Wagner, T, Wodrich, M.D, Ataka, K, Bill, E, Ermler, U, Hu, X, Shima, S. | | Deposit date: | 2018-08-07 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The atomic-resolution crystal structure of activated [Fe]-hydrogenase
Nat Catal, 2019
|
|
6HUY
 
 | | HmdII from Desulfurobacterium thermolithotrophum reconstitued with Fe-guanylylpyridinol (FeGP) cofactor and co-crystallized with methenyl-tetrahydrofolate form A | | Descriptor: | 5,10-Methenyltetrahydrofolate, Coenzyme F420-dependent N(5),N(10)-methenyltetrahydromethanopterin reductase-related protein, DIMETHYL SULFOXIDE, ... | | Authors: | Watanabe, T, Wagner, T, Huang, G, Kahnt, J, Ataka, K, Ermler, U, Shima, S. | | Deposit date: | 2018-10-09 | | Release date: | 2019-01-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Bacterial [Fe]-Hydrogenase Paralog HmdII Uses Tetrahydrofolate Derivatives as Substrates.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
6HUX
 
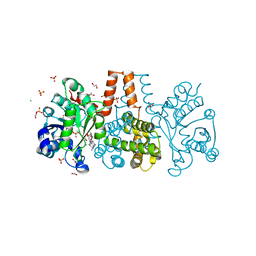 | | HmdII from Methanocaldococcus jannaschii reconstitued with Fe-guanylylpyridinol (FeGP) cofactor and co-crystallized with methenyl-tetrahydromethanopterin at 2.5 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 1-{4-[(6S,6aR,7R)-3-amino-6,7-dimethyl-1-oxo-1,2,5,6,6a,7-hexahydro-8H-imidazo[1,5-f]pteridin-10-ium-8-yl]phenyl}-1-deoxy-5-O-{5-O-[(S)-{[(1S)-1,3-dicarboxypropyl]oxy}(hydroxy)phosphoryl]-alpha-D-ribofuranosyl}-D-ribitol, ACETATE ION, ... | | Authors: | Watanabe, T, Wagner, T, Huang, G, Kahnt, J, Ataka, K, Ermler, U, Shima, S. | | Deposit date: | 2018-10-09 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Bacterial [Fe]-Hydrogenase Paralog HmdII Uses Tetrahydrofolate Derivatives as Substrates.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
6HAE
 
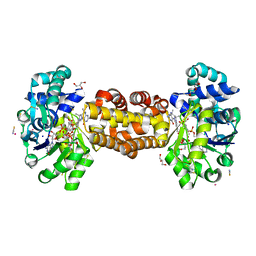 | | Crystal structure of [Fe]-hydrogenase (Hmd) from Methanococcus aeolicus in complex with FeGP cofactor and methenyl-tetrahydromethanopterin (close form B) | | Descriptor: | 1-{4-[(6S,6aR,7R)-3-amino-6,7-dimethyl-1-oxo-1,2,5,6,6a,7-hexahydro-8H-imidazo[1,5-f]pteridin-10-ium-8-yl]phenyl}-1-deoxy-5-O-{5-O-[(S)-{[(1S)-1,3-dicarboxypropyl]oxy}(hydroxy)phosphoryl]-alpha-D-ribofuranosyl}-D-ribitol, 5,10-methenyltetrahydromethanopterin hydrogenase, CHLORIDE ION, ... | | Authors: | Huang, G, Wagner, T, Wodrich, M.D, Ataka, K, Bill, E, Ermler, U, Hu, X, Shima, S. | | Deposit date: | 2018-08-07 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The atomic-resolution crystal structure of activated [Fe]-hydrogenase
Nat Catal, 2019
|
|
6HAV
 
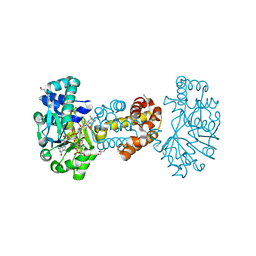 | | Crystal structure of [Fe]-hydrogenase (Hmd) from Methanococcus aeolicus in complex with FeGP and methenyl-tetrahydromethanopterin (close form A) at 1.06 A resolution | | Descriptor: | 1-{4-[(6S,6aR,7R)-3-amino-6,7-dimethyl-1-oxo-1,2,5,6,6a,7-hexahydro-8H-imidazo[1,5-f]pteridin-10-ium-8-yl]phenyl}-1-deoxy-5-O-{5-O-[(S)-{[(1S)-1,3-dicarboxypropyl]oxy}(hydroxy)phosphoryl]-alpha-D-ribofuranosyl}-D-ribitol, 5,10-methenyltetrahydromethanopterin hydrogenase, GLYCEROL, ... | | Authors: | Huang, G, Wagner, T, Wodrich, M.D, Ataka, K, Bill, E, Ermler, U, Hu, X, Shima, S. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | The atomic-resolution crystal structure of activated [Fe]-hydrogenase
Nat Catal, 2019
|
|
6EUN
 
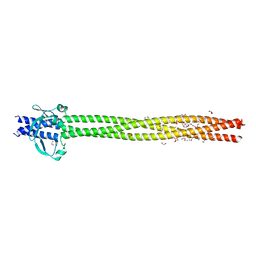 | | Crystal structure of Neisseria meningitidis vaccine antigen NadA variant 3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, ... | | Authors: | Dello Iacono, L, Liguori, A, Malito, E, Bottomley, M.J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | NadA3 Structures Reveal Undecad Coiled Coils and LOX1 Binding Regions Competed by Meningococcus B Vaccine-Elicited Human Antibodies.
MBio, 9, 2018
|
|
6EUP
 
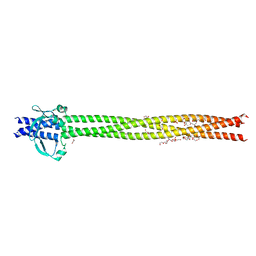 | | Crystal structure of Neisseria meningitidis NadA variant 3 double mutant A33I-I38L | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Adhesin A, ... | | Authors: | Dello Iacono, L, Liguori, A, Malito, E, Bottomley, M.J. | | Deposit date: | 2017-10-31 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | NadA3 Structures Reveal Undecad Coiled Coils and LOX1 Binding Regions Competed by Meningococcus B Vaccine-Elicited Human Antibodies.
MBio, 9, 2018
|
|
6TJR
 
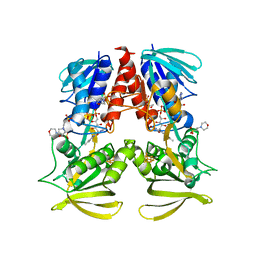 | | Structure of HdrA-like subunit from Hyphomicrobium denitrificans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Fumarate reductase/succinate dehydrogenase flavoprotein domain protein, ... | | Authors: | Kayastha, K, Ermler, U, Dahl, C. | | Deposit date: | 2019-11-26 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural and spectroscopic characterization of a HdrA-like subunit from Hyphomicrobium denitrificans.
Febs J., 288, 2021
|
|
6TLK
 
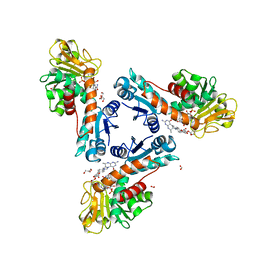 | | Structure of methylene-tetrahydromethanopterin dehydrogenase from Methylorubrum extorquens AM1 in an open conformation containing NADP+ and methylene-H4MPT | | Descriptor: | 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, Bifunctional protein MdtA, CHLORIDE ION, ... | | Authors: | Wagner, T, Huang, G, Demmer, U, Warkentin, E, Ermler, U, Shima, S. | | Deposit date: | 2019-12-02 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Hydride Transfer Process in NADP-dependent Methylene-tetrahydromethanopterin Dehydrogenase.
J.Mol.Biol., 432, 2020
|
|
6TM3
 
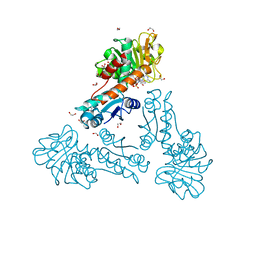 | | Structure of methylene-tetrahydromethanopterin dehydrogenase from Methylorubrum extorquens AM1 in a close conformation containing NADP+ and methylene-H4MPT | | Descriptor: | 1,2-ETHANEDIOL, 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, Bifunctional protein MdtA, ... | | Authors: | Wagner, T, Huang, G, Demmer, U, Warkentin, E, Ermler, U, Shima, S. | | Deposit date: | 2019-12-03 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | The Hydride Transfer Process in NADP-dependent Methylene-tetrahydromethanopterin Dehydrogenase.
J.Mol.Biol., 432, 2020
|
|
6QKR
 
 | |
