9ATE
 
 | | Crystal structure of MERS 3CL protease in complex with a methylbicyclo[2.2.1]heptane 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-{[N-({[(2S)-1-{[(1S,2S,4R)-bicyclo[2.2.1]heptan-2-yl]methyl}-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(2S)-1-{[(1S,2S,4R)-bicyclo[2.2.1]heptan-2-yl]methyl}-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9ATD
 
 | | Crystal structure of MERS 3CL protease in complex with a ethylcyclohexyl 2-pyrrolidone inhibitor (S-enantiomer) inhibitor | | Descriptor: | (1R,2S)-2-{[N-({[(2S)-1-(2-cyclohexylethyl)-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(2S)-1-(2-cyclohexylethyl)-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
8XGA
 
 | |
9ATF
 
 | | Crystal structure of MERS 3CL protease in complex with a 1-methyl-4,4-difluorocyclohexyl 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-({N-[({(2S)-1-[(4,4-difluorocyclohexyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[({(2S)-1-[(4,4-difluorocyclohexyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9ATT
 
 | | Crystal structure of MERS 3CL protease in complex with a methylcyclohexyl 2-pyrrolidone inhibitor (R-enantiomer) | | Descriptor: | (1R,2S)-2-{[N-({[(2R)-1-(cyclohexylmethyl)-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(2R)-1-(cyclohexylmethyl)-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-27 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
8WKY
 
 | | Crystal structure of the Melanocortin-4 Receptor (MC4R) in complex with S25 | | Descriptor: | CALCIUM ION, Melanocortin receptor 4, N-(2-aminoethyl)-5-(2-{[4-(morpholin-4-yl)pyridin-2-yl]amino}-1,3-thiazol-5-yl)pyridine-3-carboxamide, ... | | Authors: | Gimenez, L.E, Martin, C, Yu, J, Hollanders, C, Hernandez, C, Dahir, N.S, Wu, Y, Yao, D, Han, G.W, Wu, L, Poorten, O.V, Lamouroux, A, Mannes, M, Tourwe, D, Zhao, S, Stevens, R.C, Cone, R.D, Ballet, S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Novel Cocrystal Structures of Peptide Antagonists Bound to the Human Melanocortin Receptor 4 Unveil Unexplored Grounds for Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
9FKX
 
 | | Crystal structure of IL-17A in complex with compound 18 | | Descriptor: | (4~{S})-1-[(~{R})-[2-[(~{S})-[4,4-bis(fluoranyl)cyclohexyl]-[[5-[1,1-bis(fluoranyl)ethyl]-1,3,4-oxadiazol-2-yl]amino]methyl]imidazo[1,2-b]pyridazin-7-yl]-cyclopropyl-methyl]-4-(trifluoromethyl)imidazolidin-2-one, Interleukin-17A | | Authors: | Rondeau, J.M, Lehmann, S, Scheufler, C. | | Deposit date: | 2024-06-04 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.048 Å) | | Cite: | Discovery and In Vivo Exploration of 1,3,4-Oxadiazole and alpha-Fluoroacrylate Containing IL-17 Inhibitors.
J.Med.Chem., 67, 2024
|
|
9ATG
 
 | | Crystal structure of MERS 3CL protease in complex with a 2,2-difluoro-5-methylbenzo[1,3]dioxole 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-({N-[({(2S)-1-[(2,2-difluoro-2H-1,3-benzodioxol-5-yl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[({(2S)-1-[(2,2-difluoro-2H-1,3-benzodioxol-5-yl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9ATA
 
 | | Crystal structure of MERS 3CL protease in complex with a phenylethyl 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[(2S)-5-oxo-1-(2-phenylethyl)pyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[(2S)-5-oxo-1-(2-phenylethyl)pyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9BJM
 
 | | Crystal Structure of Inhibitor 5c in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 1'-{[5-chloro-1-(4,4,4-trifluorobutyl)-1H-1,3-benzimidazol-2-yl]methyl}-1-(methanesulfonyl)spiro[azetidine-3,3'-indol]-2'(1'H)-one, Prefusion RSV F (DS-CAV1),Envelope glycoprotein | | Authors: | Shaffer, P.L, Milligan, C, Abeywickrema, P. | | Deposit date: | 2024-04-25 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Spiro-Azetidine Oxindoles as Long-Acting Injectables for Pre-Exposure Prophylaxis against Respiratory Syncytial Virus Infections.
J.Med.Chem., 67, 2024
|
|
9ATS
 
 | | Crystal structure of MERS 3CL protease in complex with a methylcyclohexyl 2-pyrrolidone inhibitor (S-enantiomer) | | Descriptor: | (1S,2S)-2-{[N-({[(2S)-1-(cyclohexylmethyl)-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-27 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
7PFZ
 
 | |
8WYG
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor 22 | | Descriptor: | 2-[[5-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-1-methyl-2-oxidanylidene-pyridin-4-yl]amino]-~{N}-(4-oxidanylcyclohexyl)ethanamide, Bromodomain-containing protein 2 | | Authors: | Xu, H, Zhao, X, Shen, H, Xu, Y, Wu, X. | | Deposit date: | 2023-10-30 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Discovery of Novel Phenoxyaryl Pyridones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with High Selectivity for the Second Bromodomain (BD2) to Potentially Treat Acute Myeloid Leukemia.
J.Med.Chem., 67, 2024
|
|
9GDL
 
 | |
9BCG
 
 | | Myeloid cell leukemia-1 (Mcl-1) complexed with compound | | Descriptor: | 7-[(4R,5S,6P)-7-chloro-10-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-4-methyl-1-oxo-6-(1,3,5-trimethyl-1H-pyrazol-4-yl)-3,4-dihydropyrazino[1,2-a]indol-2(1H)-yl]-4,5-dimethoxy-1-methyl-1H-indole-2-carboxylic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhao, B, Fesik, S.W. | | Deposit date: | 2024-04-09 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Discovery of a Myeloid Cell Leukemia 1 (Mcl-1) Inhibitor That Demonstrates Potent In Vivo Activities in Mouse Models of Hematological and Solid Tumors.
J.Med.Chem., 67, 2024
|
|
8XGV
 
 | | Optimization Efforts for Identification of Novel Highly Potent Keap1-Nrf2 Protein-Protein Interaction (PPI) Inhibitors | | Descriptor: | (2~{R},3~{S})-3-[[(2~{S})-2-[4-[(3-ethoxypyridin-2-yl)methyl]phenyl]-2-fluoranyl-ethanoyl]amino]-2-methyl-3-(4-methylphenyl)propanoic acid, ACETATE ION, Kelch-like ECH-associated protein 1, ... | | Authors: | Otake, K, Hara, Y, Ubukata, M, Inoue, M, Nagahashi, N, Motoda, D, Ogawa, N, Hantani, Y, Hantani, R, Adachi, T, Nomura, A, Yamaguchi, K, Maekawa, M, Mamada, H, Motomura, T, Sato, M, Harada, K. | | Deposit date: | 2023-12-15 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Optimization Efforts for Identification of Novel Highly Potent Keap1-Nrf2 Protein-Protein Interaction Inhibitors.
J.Med.Chem., 67, 2024
|
|
7KXT
 
 | | Crystal structure of human EED | | Descriptor: | 1-[(4-fluorophenyl)methyl]-N-{1-[2-(4-methoxyphenyl)ethyl]piperidin-4-yl}-1H-benzimidazol-2-amine, Polycomb protein EED, UNKNOWN ATOM OR ION | | Authors: | Zhu, L, Dong, A, Du, D, Liu, Y, Luo, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-04 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Guided Development of Small-Molecule PRC2 Inhibitors Targeting EZH2-EED Interaction.
J.Med.Chem., 64, 2021
|
|
9C9V
 
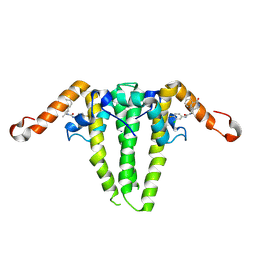 | | HBV capsid with compound 3i | | Descriptor: | Capsid protein, N'-(3-chloro-4-fluorophenyl)-N-(2-methylpropyl)-N-[(1R)-1-(1-oxo-1,2-dihydroisoquinolin-4-yl)ethyl]urea | | Authors: | Olland, A.M, Suto, R.K, Fontano, E, Colussi, T. | | Deposit date: | 2024-06-16 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Rational Design, Synthesis, and Structure-Activity Relationship of a Novel Isoquinolinone-Based Series of HBV Capsid Assembly Modulators Leading to the Identification of Clinical Candidate AB-836.
J.Med.Chem., 67, 2024
|
|
8SIX
 
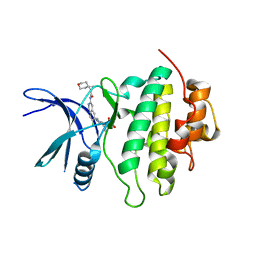 | | Structure of Compound 13 bound to the CHK1 10-point mutant | | Descriptor: | (1S)-N-(7-chloro-6-{4-[(3R,4R)-4-hydroxy-3-methyloxolan-3-yl]piperazin-1-yl}isoquinolin-3-yl)-6-oxaspiro[2.5]octane-1-carboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2023-04-17 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of MK-1468: A Potent, Kinome-Selective, Brain-Penetrant Amidoisoquinoline LRRK2 Inhibitor for the Potential Treatment of Parkinson's Disease.
J.Med.Chem., 66, 2023
|
|
8SIW
 
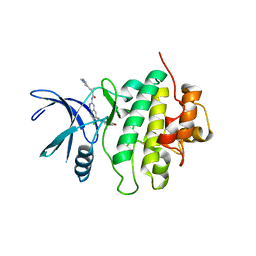 | | Structure of Compound 5 bound to the CHK1 10-point mutant | | Descriptor: | (1S,2S)-N-(7-chloro-6-{1-[(3R,4R)-4-hydroxy-3-methyloxolan-3-yl]piperidin-4-yl}isoquinolin-3-yl)-2-(1-methyl-1H-pyrazol-4-yl)cyclopropane-1-carboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2023-04-17 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.877 Å) | | Cite: | Discovery of MK-1468: A Potent, Kinome-Selective, Brain-Penetrant Amidoisoquinoline LRRK2 Inhibitor for the Potential Treatment of Parkinson's Disease.
J.Med.Chem., 66, 2023
|
|
8SIV
 
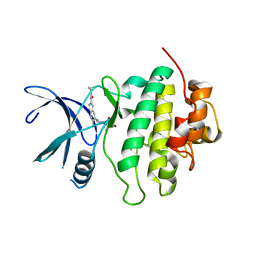 | | Structure of Compound 2 bound to the CHK1 10-point mutant | | Descriptor: | N-(7-chloro-6-{1-[(3R,4R)-4-hydroxy-3-methyloxolan-3-yl]piperidin-4-yl}isoquinolin-3-yl)cyclopropanecarboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2023-04-17 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | Discovery of MK-1468: A Potent, Kinome-Selective, Brain-Penetrant Amidoisoquinoline LRRK2 Inhibitor for the Potential Treatment of Parkinson's Disease.
J.Med.Chem., 66, 2023
|
|
6TD5
 
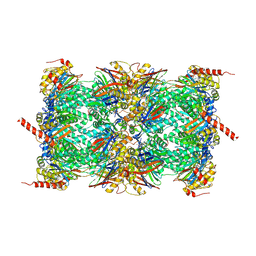 | | Leishmania tarentolae proteasome 20S subunit complexed with LXE408 and bortezomib | | Descriptor: | N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, Proteasome endopeptidase complex, Proteasome subunit alpha type, ... | | Authors: | Srinivas, H. | | Deposit date: | 2019-11-07 | | Release date: | 2020-08-26 | | Last modified: | 2020-10-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Discovery and Characterization of Clinical Candidate LXE408 as a Kinetoplastid-Selective Proteasome Inhibitor for the Treatment of Leishmaniases.
J.Med.Chem., 63, 2020
|
|
7SSB
 
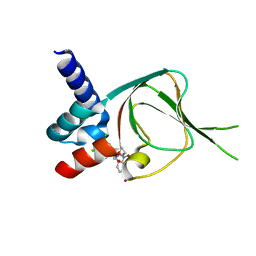 | | Co-structure of PKG1 regulatory domain with compound 33 | | Descriptor: | 4-({(2S,3S)-3-[(1S)-1-(3,5-dichlorophenyl)-2-hydroxyethoxy]-2-phenylpiperidin-1-yl}methyl)-3-nitrobenzoic acid, cGMP-dependent protein kinase 1 | | Authors: | Fischmann, T.O. | | Deposit date: | 2021-11-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Optimization and Mechanistic Investigations of Novel Allosteric Activators of PKG1 alpha.
J.Med.Chem., 65, 2022
|
|
6TPE
 
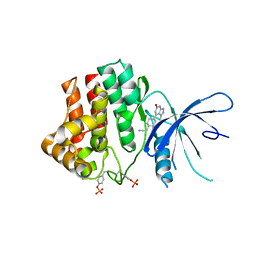 | | Fragment-based discovery of pyrazolopyridones as JAK1 inhibitors with excellent subtype selectivity | | Descriptor: | 2-[4-(3-methyl-6-oxidanylidene-1,7-dihydropyrazolo[3,4-b]pyridin-4-yl)cyclohexyl]ethanenitrile, Tyrosine-protein kinase JAK1 | | Authors: | Hansen, B.B, Jepsen, T.H, Larsen, M, Sindet, R, Vifian, T, Burhardt, M.N, Larsen, J, Seitzberg, J.G, Carnerup, M.A, Jerre, A, Molck, C, Rai, S, Nasipireddy, V.R, Jestel, A, Lammens, A, Ritzen, A. | | Deposit date: | 2019-12-13 | | Release date: | 2020-06-10 | | Last modified: | 2020-07-22 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Fragment-Based Discovery of Pyrazolopyridones as JAK1 Inhibitors with Excellent Subtype Selectivity.
J.Med.Chem., 63, 2020
|
|
7L0K
 
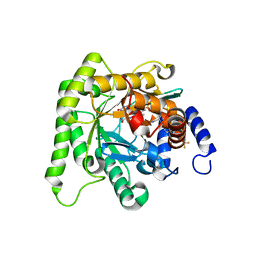 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM784 (3-(1-(3-methyl-4-((6-(trifluoromethyl)pyridin-3-yl)methyl)-1H-pyrrole-2-carboxamido)ethyl)-1H-pyrazole-5-carboxamide) | | Descriptor: | 3-{(1R)-1-[(3-methyl-4-{[6-(trifluoromethyl)pyridin-3-yl]methyl}-1H-pyrrole-2-carbonyl)amino]ethyl}-1H-pyrazole-5-carboxamide, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-11 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
