6DMW
 
 | |
4DS7
 
 | |
2KZ2
 
 | | Calmodulin, C-terminal domain, F92E mutant | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Korendovych, I, Kulp, D, Wu, Y, Cheng, H, Roder, H, DeGrado, W. | | Deposit date: | 2010-06-10 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Design of a switchable eliminase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5OEO
 
 | | Solution structure of the complex of TRPV5(655-725) with a Calmodulin E32Q/E68Q double mutant | | Descriptor: | CALCIUM ION, Calmodulin-1, Transient receptor potential cation channel subfamily V member 5 | | Authors: | Vuister, G.W, Bokhovchuk, F.M, Bate, N, Kovalevskaya, N, Goult, B.T, Spronk, C.A.E.M. | | Deposit date: | 2017-07-09 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structural Basis of Calcium-Dependent Inactivation of the Transient Receptor Potential Vanilloid 5 Channel.
Biochemistry, 57, 2018
|
|
4AQR
 
 | | Crystal structure of calmodulin in complex with the regulatory domain of a plasma-membrane Ca2+-ATPase | | Descriptor: | CALCIUM ION, CALCIUM-TRANSPORTING ATPASE 8, PLASMA MEMBRANE-TYPE, ... | | Authors: | Tidow, H, Poulsen, L.R, Andreeva, A, Hein, K.L, Palmgren, M.G, Nissen, P. | | Deposit date: | 2012-04-19 | | Release date: | 2012-10-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Bimodular Mechanism of Calcium Control in Eukaryotes
Nature, 491, 2012
|
|
5DBR
 
 | |
5L0O
 
 | |
2XZS
 
 | | Death associated protein kinase 1 residues 1-312 | | Descriptor: | DEATH ASSOCIATED KINASE 1, MAGNESIUM ION | | Authors: | Yumerefendi, H, Mas, P.J, Dordevic, N, McCarthy, A.A, Hart, D.J. | | Deposit date: | 2010-11-29 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Death-Associated Protein Kinase Activity is Regulated by Coupled Calcium/Calmodulin Binding to Two Distinct Sites.
Structure, 24, 2016
|
|
6EWW
 
 | |
5K7L
 
 | |
8KA0
 
 | | Crystal structure of Vibrio vulnificus RID-dependent transforming NADase domain (RDTND)/calmodulin-binding domain of Rho inactivation domain (RID-CBD) complexed with Ca2+-bound calmodulin and a nicotinamide adenine dinucleotide (NAD+) | | Descriptor: | CALCIUM ION, Calmodulin-2, GLYCEROL, ... | | Authors: | Lee, Y, Choi, S, Hwang, J, Kim, M.H. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Dissemination of pathogenic bacteria is reinforced by a MARTX toxin effector duet.
Nat Commun, 15, 2024
|
|
8K9Z
 
 | | Crystal structure of Vibrio vulnificus RID-dependent transforming NADase domain (RDTND)/calmodulin-binding domain of Rho inactivation domain (RID-CBD) complexed with Ca2+-bound calmodulin | | Descriptor: | CALCIUM ION, Calmodulin-2, RDTND-RID CBD | | Authors: | Lee, Y, Choi, S, Hwang, J, Kim, M.H. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dissemination of pathogenic bacteria is reinforced by a MARTX toxin effector duet.
Nat Commun, 15, 2024
|
|
4BYF
 
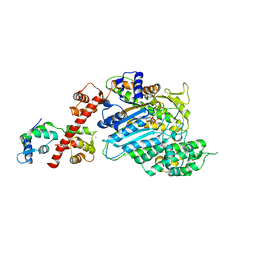 | | Crystal structure of human Myosin 1c in complex with calmodulin in the pre-power stroke state | | Descriptor: | ADP ORTHOVANADATE, CALMODULIN, MAGNESIUM ION, ... | | Authors: | Munnich, S, Taft, M.H, Pathan-Chhatbar, S, Manstein, D.J. | | Deposit date: | 2013-07-19 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal Structure of Human Myosin 1C-the Motor in Glut4 Exocytosis: Implications for Ca(2+) Regulation and 14-3-3 Binding.
J.Mol.Biol., 426, 2014
|
|
7OAJ
 
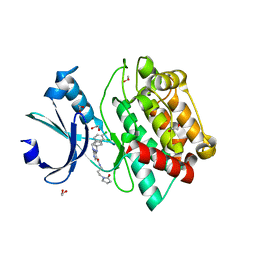 | | Crystal structure of pseudokinase CASK in complex with compound 7 | | Descriptor: | 1,2-ETHANEDIOL, 4-(cyclopentylamino)-2-[(3,4-dichlorophenyl)methylamino]-N-[3-(2-oxidanylidenepyrrolidin-1-yl)propyl]pyrimidine-5-carboxamide, Peripheral plasma membrane protein CASK | | Authors: | Chaikuad, A, Russ, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Design and Development of a Chemical Probe for Pseudokinase Ca 2+ /calmodulin-Dependent Ser/Thr Kinase.
J.Med.Chem., 64, 2021
|
|
7OAI
 
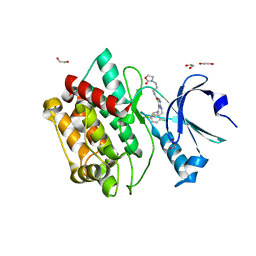 | | Crystal structure of pseudokinase CASK in complex with PFE-PKIS12 | | Descriptor: | 1,2-ETHANEDIOL, 4-(Cyclopentylamino)-2-[(2,5-dichlorophenyl)methylamino]-N-[3-(2-oxo-1,3-oxazolidin-3-yl)propyl]pyrimidine-5-carboxamide, Peripheral plasma membrane protein CASK | | Authors: | Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Development of a Chemical Probe for Pseudokinase Ca 2+ /calmodulin-Dependent Ser/Thr Kinase.
J.Med.Chem., 64, 2021
|
|
7OAK
 
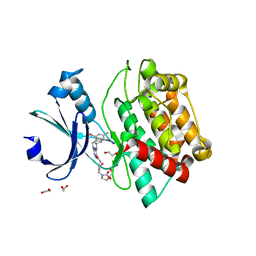 | | Crystal structure of pseudokinase CASK in complex with compound 26 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[2,5-bis(bromanyl)-4-methyl-phenyl]methylamino]-4-(cyclopentylamino)-N-[3-(2-oxidanylidene-1,3-oxazolidin-3-yl)propyl]pyrimidine-5-carboxamide, Peripheral plasma membrane protein CASK | | Authors: | Chaikuad, A, Russ, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Design and Development of a Chemical Probe for Pseudokinase Ca 2+ /calmodulin-Dependent Ser/Thr Kinase.
J.Med.Chem., 64, 2021
|
|
7OAL
 
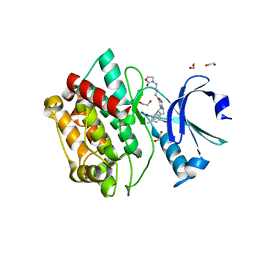 | | Crystal structure of pseudokinase CASK in complex with compound 25 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[2,5-bis(bromanyl)-4-methyl-phenyl]methylamino]-4-(cyclohexylamino)-N-[3-(2-oxidanylidene-1,3-oxazolidin-3-yl)propyl]pyrimidine-5-carboxamide, Peripheral plasma membrane protein CASK | | Authors: | Chaikuad, A, Russ, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Design and Development of a Chemical Probe for Pseudokinase Ca 2+ /calmodulin-Dependent Ser/Thr Kinase.
J.Med.Chem., 64, 2021
|
|
6OF9
 
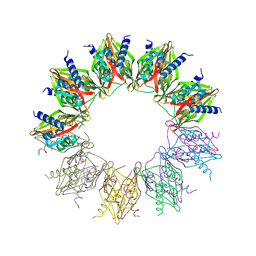 | |
6SZ5
 
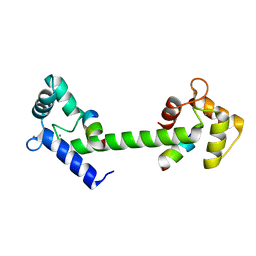 | |
8QMP
 
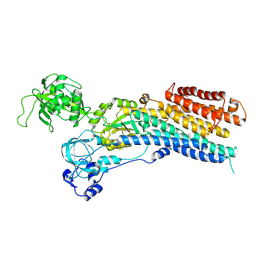 | | Structure of the E2 Beryllium Fluoride Complex of the Autoinhibited Calcium ATPase ACA8 | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Calcium-transporting ATPase 8, plasma membrane-type, ... | | Authors: | Thirup Larsen, S, Karlsen Dannersoe, J, Nissen, P. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conserved N-terminal Regulation of the ACA8 Calcium Pump with Two Calmodulin Binding Sites.
J.Mol.Biol., 436, 2024
|
|
5SY1
 
 | | Structure of the STRA6 receptor for retinol uptake in complex with calmodulin | | Descriptor: | CALCIUM ION, CHOLESTEROL, Calmodulin, ... | | Authors: | Clarke, O.B, Chen, Y, Mancia, F. | | Deposit date: | 2016-08-10 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the STRA6 receptor for retinol uptake.
Science, 353, 2016
|
|
5IG4
 
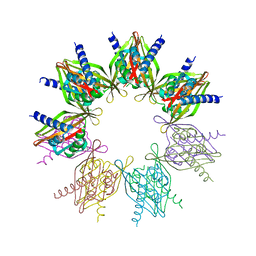 | | Crystal structure of N. vectensis CaMKII-A hub | | Descriptor: | GLYCEROL, Predicted protein | | Authors: | Bhattacharyya, M, Pappireddi, N, Gee, C.L, Barros, T, Kuriyan, J. | | Deposit date: | 2016-02-26 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular mechanism of activation-triggered subunit exchange in Ca(2+)/calmodulin-dependent protein kinase II.
Elife, 5, 2016
|
|
5IG0
 
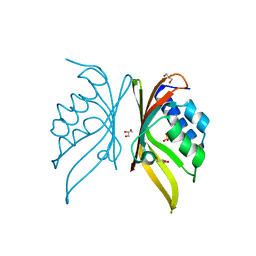 | | Crystal structure of S. rosetta CaMKII hub | | Descriptor: | CAMK/CAMK2 protein kinase, GLYCEROL, SULFATE ION | | Authors: | Bhattacharyya, M, Gee, C.L, Barros, T, Kuriyan, J. | | Deposit date: | 2016-02-26 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular mechanism of activation-triggered subunit exchange in Ca(2+)/calmodulin-dependent protein kinase II.
Elife, 5, 2016
|
|
5IG5
 
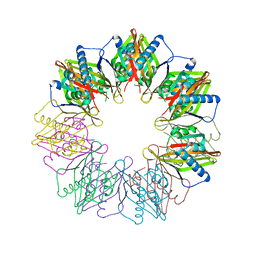 | |
6OQQ
 
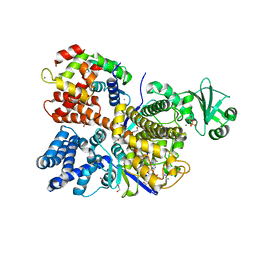 | | Legionella pneumophila SidJ/Saccharomyces cerevisiae calmodulin complex | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, CALCIUM ION, ... | | Authors: | Tomchick, D.R, Tagliabracci, V.S, Black, M, Osinski, A. | | Deposit date: | 2019-04-28 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Bacterial pseudokinase catalyzes protein polyglutamylation to inhibit the SidE-family ubiquitin ligases.
Science, 364, 2019
|
|
