4JMH
 
 | |
1CZD
 
 | | CRYSTAL STRUCTURE OF THE PROCESSIVITY CLAMP GP45 FROM BACTERIOPHAGE T4 | | Descriptor: | DNA POLYMERASE ACCESSORY PROTEIN G45 | | Authors: | Moarefi, I, Jeruzalmi, D, Turner, J, O'Donnell, M, Kuriyan, J. | | Deposit date: | 1999-09-02 | | Release date: | 2000-03-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the DNA polymerase processivity factor of T4 bacteriophage.
J.Mol.Biol., 296, 2000
|
|
2NTI
 
 | | Crystal structure of PCNA123 heterotrimer. | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, BROMIDE ION, DNA polymerase sliding clamp A, ... | | Authors: | Hlinkova, V, Ling, H. | | Deposit date: | 2006-11-07 | | Release date: | 2007-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of monomeric, dimeric and trimeric PCNA: PCNA-ring assembly and opening.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
8UII
 
 | |
1RWZ
 
 | | Crystal Structure of Proliferating Cell Nuclear Antigen (PCNA) from A. fulgidus | | Descriptor: | DNA polymerase sliding clamp | | Authors: | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | Deposit date: | 2003-12-17 | | Release date: | 2004-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
1RXM
 
 | | C-terminal region of FEN-1 bound to A. fulgidus PCNA | | Descriptor: | DNA polymerase sliding clamp, consensus FEN-1 peptide | | Authors: | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | Deposit date: | 2003-12-18 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
4MJR
 
 | | E. coli sliding clamp in complex with (S)-Carprofen | | Descriptor: | (2S)-2-(6-chloro-9H-carbazol-2-yl)propanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-09-04 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | DNA replication is the target for the antibacterial effects of nonsteroidal anti-inflammatory drugs.
Chem.Biol., 21, 2014
|
|
4MJP
 
 | | E. coli sliding clamp in complex with (R)-Vedaprofen | | Descriptor: | (2R)-2-(4-cyclohexylnaphthalen-1-yl)propanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-09-04 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | DNA replication is the target for the antibacterial effects of nonsteroidal anti-inflammatory drugs.
Chem.Biol., 21, 2014
|
|
4MJQ
 
 | | E. coli sliding clamp in complex with Bromfenac | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-09-04 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | DNA replication is the target for the antibacterial effects of nonsteroidal anti-inflammatory drugs.
Chem.Biol., 21, 2014
|
|
7TFI
 
 | | Atomic model of the S. cerevisiae clamp-clamp loader complex PCNA-RFC bound to DNA with an open clamp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2022-01-06 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Cryo-EM structures reveal that RFC recognizes both the 3'- and 5'-DNA ends to load PCNA onto gaps for DNA repair.
Elife, 11, 2022
|
|
7TFJ
 
 | | Atomic model of S. cerevisiae clamp-clamp loader complex PCNA-RFC bound to DNA with a closed clamp ring | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2022-01-06 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures reveal that RFC recognizes both the 3'- and 5'-DNA ends to load PCNA onto gaps for DNA repair.
Elife, 11, 2022
|
|
8UI9
 
 | |
8UI8
 
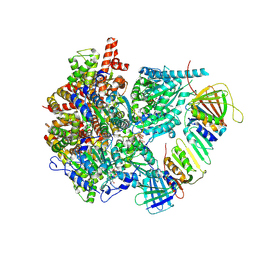 | |
2HIK
 
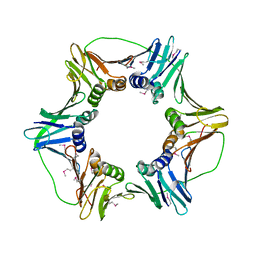 | | heterotrimeric PCNA sliding clamp | | Descriptor: | PCNA1 (SSO0397), PCNA2 (SSO1047), PCNA3 (SSO0405) | | Authors: | Pascal, J.M, Tsodikov, O.V, Ellenberger, T. | | Deposit date: | 2006-06-29 | | Release date: | 2006-11-07 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A Flexible Interface between DNA Ligase and PCNA Supports Conformational Switching and Efficient Ligation of DNA.
Mol.Cell, 24, 2006
|
|
2IO4
 
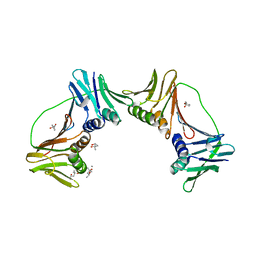 | | Crystal structure of PCNA12 dimer from Sulfolobus solfataricus. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, DNA polymerase sliding clamp B, ... | | Authors: | Hlinkova, V, Ling, H. | | Deposit date: | 2006-10-09 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of monomeric, dimeric and trimeric PCNA: PCNA-ring assembly and opening.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
7TFH
 
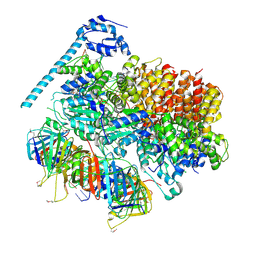 | | Atomic model of the S. cerevisiae clamp-clamp loader complex PCNA-RFC bound to two DNA molecules, one at the 5'-recessed end and the other at the 3'-recessed end | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2022-01-06 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Cryo-EM structures reveal that RFC recognizes both the 3'- and 5'-DNA ends to load PCNA onto gaps for DNA repair.
Elife, 11, 2022
|
|
5KL1
 
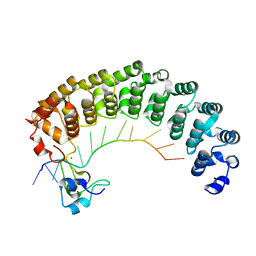 | | Crystal structure of the Pumilio-Nos-hunchback RNA complex | | Descriptor: | Maternal protein pumilio, Protein nanos, RNA (5'-R(*AP*AP*AP*UP*UP*GP*UP*AP*CP*AP*UP*A)-3'), ... | | Authors: | Qiu, C, Hall, T.M.T. | | Deposit date: | 2016-06-23 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.701 Å) | | Cite: | Drosophila Nanos acts as a molecular clamp that modulates the RNA-binding and repression activities of Pumilio.
Elife, 5, 2016
|
|
5KL8
 
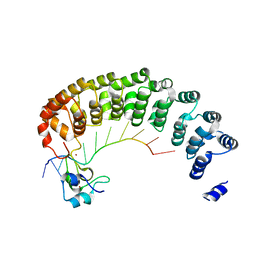 | | Crystal structure of the Pumilio-Nos-CyclinB RNA complex | | Descriptor: | Maternal protein pumilio, Protein nanos, RNA (5'-R(*UP*AP*UP*UP*UP*GP*UP*AP*AP*UP*U)-3'), ... | | Authors: | Qiu, C, Hall, T.M.T. | | Deposit date: | 2016-06-23 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Drosophila Nanos acts as a molecular clamp that modulates the RNA-binding and repression activities of Pumilio.
Elife, 5, 2016
|
|
2IZO
 
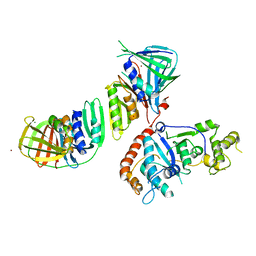 | | Structure of an Archaeal PCNA1-PCNA2-FEN1 Complex | | Descriptor: | DNA POLYMERASE SLIDING CLAMP B, DNA POLYMERASE SLIDING CLAMP C, FLAP STRUCTURE-SPECIFIC ENDONUCLEASE, ... | | Authors: | Dore, A.S, Kilkenny, M.L, Roe, S.M, Pearl, L.H. | | Deposit date: | 2006-07-25 | | Release date: | 2006-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of an Archaeal PCNA1-PCNA2-Fen1 Complex: Elucidating PCNA Subunit and Client Enzyme Specificity.
Nucleic Acids Res., 34, 2006
|
|
3Q4J
 
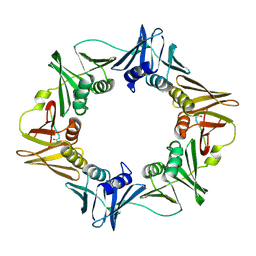 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, peptide ligand | | Authors: | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | Deposit date: | 2010-12-23 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
3Q4K
 
 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, peptide ligand | | Authors: | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | Deposit date: | 2010-12-23 | | Release date: | 2011-12-28 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
3Q4L
 
 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, SODIUM ION, peptide ligand | | Authors: | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | Deposit date: | 2010-12-23 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
2IX2
 
 | | Crystal structure of the heterotrimeric PCNA from Sulfolobus solfataricus | | Descriptor: | DNA POLYMERASE SLIDING CLAMP A, DNA POLYMERASE SLIDING CLAMP B, DNA POLYMERASE SLIDING CLAMP C | | Authors: | Williams, G.J, Johnson, K, McMahon, S.A, Carter, L, Oke, M, Liu, H, Taylor, G.L, White, M.F, Naismith, J.H. | | Deposit date: | 2006-07-05 | | Release date: | 2006-10-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Heterotrimeric PCNA from Sulfolobus Solfataricus.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2HII
 
 | | heterotrimeric PCNA sliding clamp | | Descriptor: | PCNA1 (SSO0397), PCNA2 (SSO1047), PCNA3 (SSO0405) | | Authors: | Pascal, J.M, Tsodikov, O.V, Ellenberger, T. | | Deposit date: | 2006-06-29 | | Release date: | 2006-11-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | A Flexible Interface between DNA Ligase and PCNA Supports Conformational Switching and Efficient Ligation of DNA.
Mol.Cell, 24, 2006
|
|
2IJX
 
 | |
