2H0R
 
 | | Structure of the Yeast Nicotinamidase Pnc1p | | Descriptor: | Nicotinamidase, ZINC ION | | Authors: | Taylor, A.B, Hu, G, Hart, P.J, McAlister-Henn, L. | | Deposit date: | 2006-05-15 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the yeast nicotinamidase Pnc1p.
Arch.Biochem.Biophys., 461, 2007
|
|
2K9D
 
 | | Solution structure of the domain X of measle phosphoprotein | | Descriptor: | Phosphoprotein | | Authors: | Gely, S, Bernard, C, Bourhis, J.M, Longhi, S, Darbon, H. | | Deposit date: | 2008-10-08 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Interaction between the C-terminal domains of N and P proteins of measles virus investigated by NMR.
Febs Lett., 583, 2009
|
|
2M41
 
 | |
2MI6
 
 | |
2MH0
 
 | |
2MDV
 
 | |
2LV2
 
 | | Solution NMR structure of C2H2-type Zinc-fingers 4 and 5 from human Insulinoma-associated protein 1 (fragment 424-497), Northeast Structural Genomics Consortium Target HR7614B | | Descriptor: | Insulinoma-associated protein 1, ZINC ION | | Authors: | Yang, Y, Ramelot, T.A, Cort, J.R, Shastry, R, Kohan, E, Janjua, H, Xiao, R, Acton, T, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-27 | | Release date: | 2012-10-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of C2H2-type Zinc-fingers 4 and 5 from human Insulinoma-associated protein 1 (fragment 424-497), Northeast Structural
Genomics Consortium Target HR7614B (CASP Target)
To be Published
|
|
7ESF
 
 | | The Crystal Structure of human MTH1 from Biortus | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL | | Authors: | Wang, F, Cheng, W, Shang, H, Wang, R, Zhang, B, Tian, F. | | Deposit date: | 2021-05-10 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Crystal Structure of human MTH1 from Biortus
To Be Published
|
|
8HLQ
 
 | |
8HKN
 
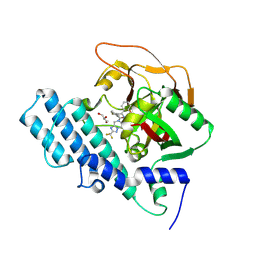 | |
8HLJ
 
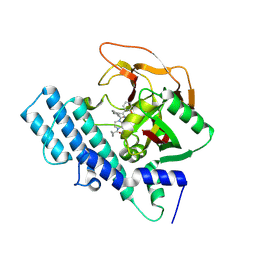 | |
8HKO
 
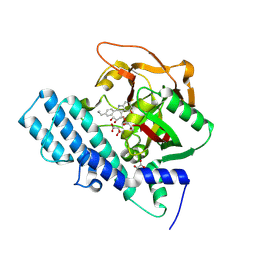 | |
8HKS
 
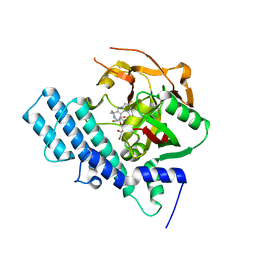 | | Mutated human ADP-ribosyltransferase 2 (PARP2) catalytic domain bound to Pamiparib(BGB-290) | | Descriptor: | (2R)-14-fluoro-2-methyl-6,9,10,19-tetrazapentacyclo[14.2.1.02,6.08,18.012,17]nonadeca-1(18),8,12(17),13,15-pentaen-11-one, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2022-11-28 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mutated human ADP-ribosyltransferase 2 (PARP2) catalytic domain bound to Pamiparib
To Be Published
|
|
8I1C
 
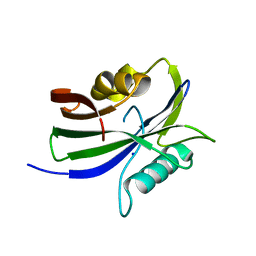 | |
8I1A
 
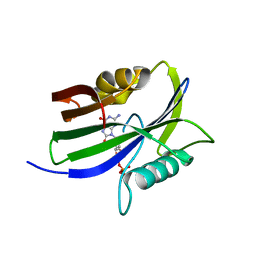 | |
8I1I
 
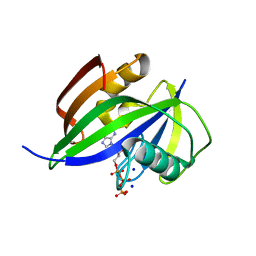 | | Crystal structure of human MTH1(G2K/D120N mutant) in complex with 2-oxo-dATP at pH 7.7 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I1D
 
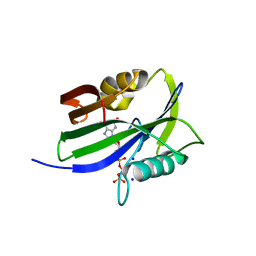 | | Crystal structure of human MTH1(G2K mutant) in complex with 2-oxo-dATP at pH 7.7 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I1E
 
 | | Crystal structure of human MTH1(G2K mutant) in complex with 2-oxo-dATP at pH 8.0 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I18
 
 | |
8I1H
 
 | |
8I1G
 
 | |
8I19
 
 | |
8I1J
 
 | | Crystal structure of human MTH1(G2K/D120N mutant) in complex with 2-oxo-dATP at pH 9.7 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I8T
 
 | |
2W99
 
 | | Crystal Structure of CDK4 in complex with a D-type cyclin | | Descriptor: | CELL DIVISION PROTEIN KINASE 4, G1/S-SPECIFIC CYCLIN-D1 | | Authors: | Day, P.J, Cleasby, A, Tickle, I.J, Reilly, M.O, Coyle, J.E, Holding, F.P, McMenamin, R.L, Yon, J, Chopra, R, Lengauer, C, Jhoti, H. | | Deposit date: | 2009-01-22 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Human Cdk4 in Complex with a D-Type Cyclin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
