2LAV
 
 | | NMR solution structure of human Vaccinia-Related Kinase 1 | | Descriptor: | Vaccinia-related kinase 1 | | Authors: | Shin, J, Yoon, H.S. | | Deposit date: | 2011-03-21 | | Release date: | 2011-05-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Human Vaccinia-related Kinase 1 (VRK1) Reveals the C-terminal Tail Essential for Its Structural Stability and Autocatalytic Activity.
J.Biol.Chem., 286, 2011
|
|
2Z0M
 
 | | Crystal structure of hypothetical ATP-dependent RNA helicase from Sulfolobus tokodaii | | Descriptor: | 337aa long hypothetical ATP-dependent RNA helicase deaD | | Authors: | Nakagawa, N, Kusano, S, Shirouzu, M, Chen, L, Fu, Z.-Q, Chrzas, J, Wang, B.-C, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of hypothetical ATP-dependent RNA helicase from Sulfolobus tokodaii
To be Published
|
|
2LQM
 
 | | Solution Structures of RadA intein from Pyrococcus horikoshii | | Descriptor: | Pho radA intein | | Authors: | Oeemig, J.S, Zhou, D, Kajander, T, Wlodawer, A, Iwai, H. | | Deposit date: | 2012-03-09 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR and Crystal Structures of the Pyrococcus horikoshii RadA Intein Guide a Strategy for Engineering a Highly Efficient and Promiscuous Intein.
J.Mol.Biol., 421, 2012
|
|
3WNT
 
 | | Multiple binding modes of benzyl isothiocyanate inhibitor complexed with Macrophage Migration Inhibitory Factor | | Descriptor: | ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor, N-benzylthioformamide, ... | | Authors: | Spencer, E.S, Dale, E.J, Gommans, A.L, Rutledge, M.T, Gamble, A.B, Smith, R.A.J, Wilbanks, S.M, Hampton, M.B, Tyndall, J.D.A. | | Deposit date: | 2013-12-16 | | Release date: | 2014-03-05 | | Last modified: | 2015-04-01 | | Method: | X-RAY DIFFRACTION (2.074 Å) | | Cite: | Multiple binding modes of isothiocyanates that inhibit macrophage migration inhibitory factor
Eur.J.Med.Chem., 93, 2015
|
|
2H94
 
 | |
2HAU
 
 | | Apo-Human Serum Transferrin (Non-Glycosylated) | | Descriptor: | CITRIC ACID, GLYCEROL, Serotransferrin | | Authors: | Wally, J, Everse, S.J. | | Deposit date: | 2006-06-13 | | Release date: | 2006-06-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of Iron-free Human Serum Transferrin Provides Insight into Inter-lobe Communication and Receptor Binding.
J.Biol.Chem., 281, 2006
|
|
2BSD
 
 | | Structure of Lactococcal Bacteriophage p2 Receptor Binding Protein | | Descriptor: | RECEPTOR BINDING PROTEIN | | Authors: | Spinelli, S, Desmyter, A, Verrips, C.T, Dehaard, H.J.W, Moineau, S, Cambillau, C. | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lactococcal Bacteriophage P2 Receptor-Binding Protein Structure Suggests a Common Ancestor Gene with Bacterial and Mammalian Viruses.
Nat.Struct.Mol.Biol., 13, 2006
|
|
3EES
 
 | |
3WNS
 
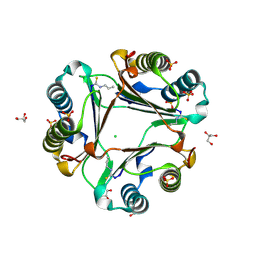 | | Allyl isothiocyanate inhibitor complexed with Macrophage Migration Inhibitory Factor | | Descriptor: | CHLORIDE ION, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Spencer, E.S, Dale, E.J, Gommans, A.L, Rutledge, M.T, Gamble, A.B, Smith, R.A.J, Wilbanks, S.M, Hampton, M.B, Tyndall, J.D.A. | | Deposit date: | 2013-12-16 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.658 Å) | | Cite: | Multiple binding modes of isothiocyanates that inhibit macrophage migration inhibitory factor
Eur.J.Med.Chem., 93, 2015
|
|
2BZW
 
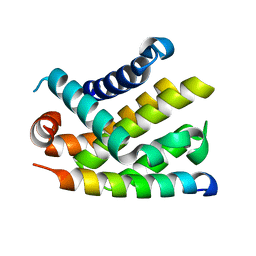 | | The crystal structure of BCL-XL in complex with full-length BAD | | Descriptor: | APOPTOSIS REGULATOR BCL-X, BCL2-ANTAGONIST OF CELL DEATH | | Authors: | Lee, K.-H, Han, W.-D, Kim, K.-J, Oh, B.-H. | | Deposit date: | 2005-08-24 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Bases for the Inhibition of Autophagy and Apoptosis by Viral Bcl-2 of Murine Gamma-Herpesvirus 68.
Plos Pathog., 4, 2008
|
|
2HKO
 
 | | Crystal structure of LSD1 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1 | | Authors: | Chen, Y, Yang, Y.T, Wang, F, Yanane, K, Zhang, Y, Lei, M. | | Deposit date: | 2006-07-05 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human histone lysine-specific demethylase 1 (LSD1).
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
7BLV
 
 | |
2HNX
 
 | | Crystal Structure of aP2 | | Descriptor: | ACETIC ACID, Fatty acid-binding protein, adipocyte, ... | | Authors: | Marr, E, Tardie, M, Carty, M, Brown Phillips, T, Qiu, X, Karam, G. | | Deposit date: | 2006-07-13 | | Release date: | 2006-11-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Expression, purification, crystallization and structure of human adipocyte lipid-binding protein (aP2).
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2Z4P
 
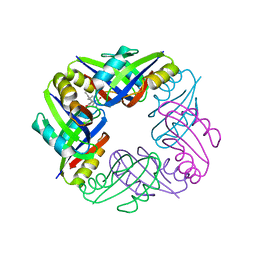 | | Crystal structure of FFRP-DM1 | | Descriptor: | 75aa long hypothetical regulatory protein AsnC, ISOLEUCINE | | Authors: | Yamada, M, Koike, H, Kudo, N, Suzuki, M. | | Deposit date: | 2007-06-21 | | Release date: | 2007-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Structural Code for Discriminating between Transcription Signals Revealed by the Feast/Famine Regulatory Protein DM1 in Complex with Ligands
Structure, 15, 2007
|
|
7BM0
 
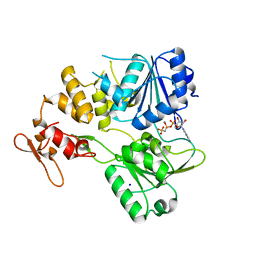 | | Crystal structure of the tick-borne encephalitis virus NS3 helicase in complex with AMPPNP | | Descriptor: | MANGANESE (II) ION, NS3 helicase domain, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Anindita, P.D, Grinkevich, P, Franta, Z. | | Deposit date: | 2021-01-19 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insight into the RNA-stimulated ATPase activity of tick-borne encephalitis virus helicase.
J.Biol.Chem., 298, 2022
|
|
2ZGY
 
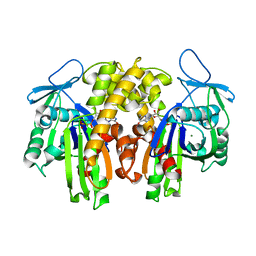 | | PARM with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Plasmid segregation protein parM | | Authors: | Popp, D, Narita, A, Oda, T, Fujisawa, T, Matsuo, H, Nitanai, Y, Iwasa, M, Maeda, K, Onishi, H, Maeda, Y. | | Deposit date: | 2008-01-30 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular structure of the ParM polymer and the mechanism leading to its nucleotide-driven dynamic instability
Embo J., 27, 2008
|
|
2LUY
 
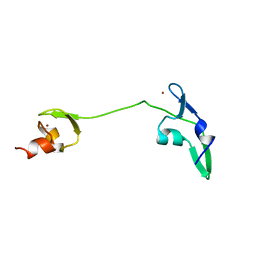 | | Solution structure of the tandem zinc finger domain of fission yeast Stc1 | | Descriptor: | Meiotic chromosome segregation protein P8B7.28c, ZINC ION | | Authors: | He, C, Shi, Y, Bayne, E, Wu, J. | | Deposit date: | 2012-06-22 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of Stc1 provides insights into the coupling of RNAi and chromatin modification.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4ETK
 
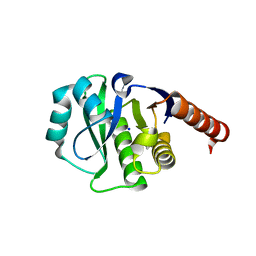 | | Crystal Structure of E6A/L130D/A155H variant of de novo designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR186 | | Descriptor: | De novo designed serine hydrolase, SODIUM ION | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
3OY3
 
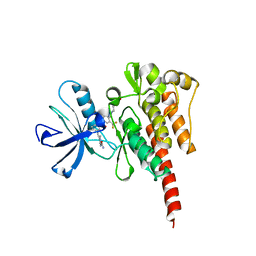 | | Crystal structure of ABL T315I mutant kinase domain bound with a DFG-out inhibitor AP24589 | | Descriptor: | 5-[(5-{[4-{[4-(2-hydroxyethyl)piperazin-1-yl]methyl}-3-(trifluoromethyl)phenyl]carbamoyl}-2-methylphenyl)ethynyl]-1-methyl-1H-imidazole-2-carboxamide, Tyrosine-protein kinase ABL1 | | Authors: | Zhou, T, Commodore, L, Huang, W.S, Wang, Y, Thomas, M, Keats, J, Xu, Q, Rivera, V, Shakespeare, W.C, Clackson, T, Dalgarno, D.C, Zhu, X. | | Deposit date: | 2010-09-22 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Mechanism of the Pan-BCR-ABL Inhibitor Ponatinib (AP24534): Lessons for Overcoming Kinase Inhibitor Resistance.
Chem.Biol.Drug Des., 77, 2011
|
|
3P1G
 
 | |
2HU2
 
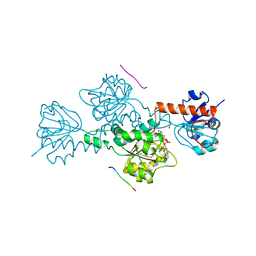 | | CTBP/BARS in ternary complex with NAD(H) and RRTGAPPAL peptide | | Descriptor: | 9-mer peptide from Zinc finger protein 217, C-terminal-binding protein 1, FORMIC ACID, ... | | Authors: | Nardini, M, Bolognesi, M, Quinlan, K.G.R, Verger, A, Francescato, P, Crossley, M. | | Deposit date: | 2006-07-26 | | Release date: | 2006-10-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Specific Recognition of ZNF217 and Other Zinc Finger Proteins at a Surface Groove of C-Terminal Binding Proteins
Mol.Cell.Biol., 26, 2006
|
|
3P6D
 
 | |
3EHW
 
 | | Human dUTPase in complex with alpha,beta-imido-dUTP and Mg2+: visualization of the full-length C-termini in all monomers and suggestion for an additional metal ion binding site | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, MAGNESIUM ION, dUTP pyrophosphatase | | Authors: | Takacs, E, Barabas, O, Vertessy, B.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-09-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human dUTPase in complex with alpha,beta-imido-dUTP and Mg2+: visualization of the full-length C-termini in all monomers and suggestion for an additional metal ion binding site
To be Published
|
|
2ZHC
 
 | | ParM filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Plasmid segregation protein parM | | Authors: | Popp, D, Narita, A, Oda, T, Fujisawa, T, Matsuo, H, Nitanai, Y, Iwasa, M, Maeda, K, Onishi, H, Maeda, Y. | | Deposit date: | 2008-02-04 | | Release date: | 2008-02-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Molecular structure of the ParM polymer and the mechanism leading to its nucleotide-driven dynamic instability
Embo J., 27, 2008
|
|
3OW8
 
 | | Crystal Structure of the WD repeat-containing protein 61 | | Descriptor: | UNKNOWN ATOM OR ION, WD repeat-containing protein 61 | | Authors: | Tempel, W, Li, Z, Chao, X, Lam, R, Wernimont, A.K, He, H, Seitova, A, Pan, P.W, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-17 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of WD40 domain proteins.
Protein Cell, 2, 2011
|
|
