8RAH
 
 | |
2FC3
 
 | | Crystal structure of the extremely thermostable Aeropyrum pernix L7Ae multifunctional protein | | Descriptor: | 50S ribosomal protein L7Ae | | Authors: | Brown II, B.A, Suryadi, J, Zhou, Z, Gupton Jr, T.B, Flowers, S.L. | | Deposit date: | 2005-12-11 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of the Aeropyrum pernix L7Ae multifunctional protein and insight into its extreme thermostability.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
7Z2R
 
 | | Differences between the GluD1 and GluD2 receptors revealed by GluD1 X-ray crystallography, binding studies and molecular dynamics | | Descriptor: | Glutamate receptor ionotropic, delta-1, SULFATE ION | | Authors: | Masternak, M, Laulumaa, S, Kastrup, J.S. | | Deposit date: | 2022-02-28 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.574 Å) | | Cite: | Differences between the GluD1 and GluD2 receptors revealed by GluD1 X-ray crystallography, binding studies and molecular dynamics.
Febs J., 290, 2023
|
|
7ZF0
 
 | | Crystal structure of UGT85B1 from Sorghum bicolor in complex with UDP and p-hydroxymandelonitrile | | Descriptor: | (2S)-HYDROXY(4-HYDROXYPHENYL)ETHANENITRILE, 1,2-ETHANEDIOL, Cyanohydrin beta-glucosyltransferase, ... | | Authors: | Putkaradze, N, Fredslund, F, Welner, D.H. | | Deposit date: | 2022-03-31 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided engineering of key amino acids in UGT85B1 controlling substrate and stereo-specificity in aromatic cyanogenic glucoside biosynthesis.
Plant J., 111, 2022
|
|
7ZER
 
 | | Crystal structure of UGT85B1 from Sorghum bicolor in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, Cyanohydrin beta-glucosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Putkaradze, N, Fredslund, F, Welner, D.H. | | Deposit date: | 2022-03-31 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure-guided engineering of key amino acids in UGT85B1 controlling substrate and stereo-specificity in aromatic cyanogenic glucoside biosynthesis.
Plant J., 111, 2022
|
|
7ZA4
 
 | | GSTF sh155 mutant | | Descriptor: | Glutathione transferase, SODIUM ION | | Authors: | Papageorgiou, A.C. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Directed Evolution of Phi Class Glutathione Transferases Involved in Multiple-Herbicide Resistance of Grass Weeds and Crops.
Int J Mol Sci, 23, 2022
|
|
7Z6T
 
 | | Aspergillus clavatus M36 protease without the propeptide | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Extracellular metalloproteinase mep, ... | | Authors: | Wilkens, C, Qiu, J, Meyer, A.S, Morth, J.P. | | Deposit date: | 2022-03-14 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Aspergillus clavatus M36 protease without the propeptide
To Be Published
|
|
7ZOS
 
 | | Class 1 Phytoglobin from Sugar beet (BvPgb1.2) | | Descriptor: | CYANIDE ION, HEXACYANOFERRATE(3-), Non-symbiotic hemoglobin class 1, ... | | Authors: | Nyblom, M, Christensen, S, Eriksson, N, Bulow, L. | | Deposit date: | 2022-04-26 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Oxidative Implications of Substituting a Conserved Cysteine Residue in Sugar Beet Phytoglobin BvPgb 1.2.
Antioxidants, 11, 2022
|
|
7ZOI
 
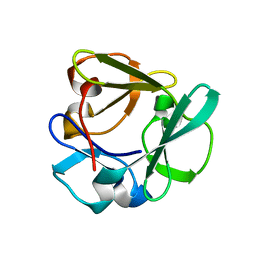 | | Carbohydrate binding domain CBM92-A from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 | | Descriptor: | Glycoside hydrolase family 18 | | Authors: | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | Deposit date: | 2022-04-25 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
7ZOO
 
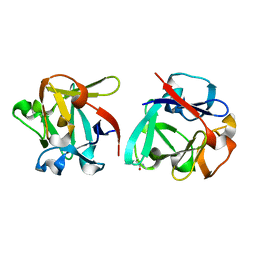 | | Carbohydrate binding domain CBM92-B from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 in complex with gentiobiose | | Descriptor: | Glycoside hydrolase family 18, beta-D-glucopyranose | | Authors: | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | Deposit date: | 2022-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
7ZON
 
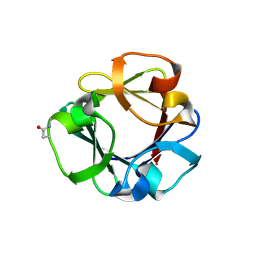 | | Carbohydrate binding domain CBM92-B from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 in complex with glucose | | Descriptor: | Glycoside hydrolase family 18, PENTAETHYLENE GLYCOL, beta-D-glucopyranose | | Authors: | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | Deposit date: | 2022-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
7ZOH
 
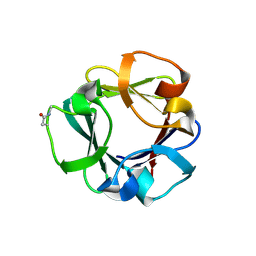 | | Carbohydrate binding domain CBM92-B from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 | | Descriptor: | Glycoside hydrolase family 18 | | Authors: | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | Deposit date: | 2022-04-25 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
7ZOP
 
 | | Carbohydrate binding domain CBM92-B from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 in complex with sophorose. | | Descriptor: | Glycoside hydrolase family 18, beta-D-glucopyranose | | Authors: | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | Deposit date: | 2022-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
7Z1U
 
 | | Biochemical implications of the substitution of a unique cysteine residue in sugar beet phytoglobin BvPgb 1.2 | | Descriptor: | Non-symbiotic hemoglobin class 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nyblom, M, Christensen, S, Leiva Eriksson, N, Bulow, L. | | Deposit date: | 2022-02-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Oxidative Implications of Substituting a Conserved Cysteine Residue in Sugar Beet Phytoglobin BvPgb 1.2.
Antioxidants, 11, 2022
|
|
7Z3Y
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH013545 | | Descriptor: | 2-[4-(3,5-dimethylpyrazol-1-yl)-2,6-bis(fluoranyl)phenyl]-~{N}-(4,5,6,7-tetrahydro-1,2-benzoxazol-3-yl)ethanamide, GLYCEROL, N-glycosylase/DNA lyase, ... | | Authors: | Scaletti, E.R, Stenmark, P. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-22 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Virtual fragment screening for DNA repair inhibitors in vast chemical space.
Nat Commun, 16, 2025
|
|
1JU8
 
 | | Solution structure of Leginsulin, a plant hormon | | Descriptor: | Leginsulin | | Authors: | Yamazaki, T, Takaoka, M, Katoh, E, Hanada, K, Sakita, M, Sakata, K, Nishiuchi, Y, Hirano, H. | | Deposit date: | 2001-08-23 | | Release date: | 2003-06-17 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | A possible physiological function and the tertiary structure of a 4-kDa peptide in legumes
EUR.J.BIOCHEM., 270, 2003
|
|
2HD5
 
 | | USP2 in complex with ubiquitin | | Descriptor: | Polyubiquitin, Ubiquitin carboxyl-terminal hydrolase 2, ZINC ION | | Authors: | Renatus, M, Kroemer, M. | | Deposit date: | 2006-06-20 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of Ubiquitin Recognition by the Deubiquitinating Protease USP2.
Structure, 14, 2006
|
|
1OZ1
 
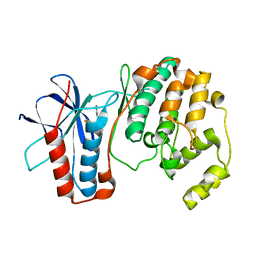 | | P38 MITOGEN-ACTIVATED KINASE IN COMPLEX WITH 4-AZAINDOLE INHIBITOR | | Descriptor: | 3-(4-FLUOROPHENYL)-2-PYRIDIN-4-YL-1H-PYRROLO[3,2-B]PYRIDIN-1-OL, Mitogen-activated protein kinase 14 | | Authors: | Lovejoy, B, Villasenor, A, Browner, M, Dunten, P. | | Deposit date: | 2003-04-07 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design and synthesis of 4-azaindoles as inhibitors of p38 MAP kinase.
J.Med.Chem., 46, 2003
|
|
7PQ2
 
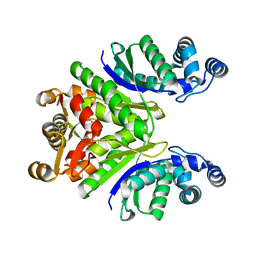 | | Crystal Structure of the Ring Nuclease 0811 from Sulfolobus islandicus (Sis0811) in its apo form | | Descriptor: | CRISPR-associated protein, APE2256 family, CRISPR Ring Nuclease | | Authors: | Molina, R, Jensen, A.L.G, Marchena-Hurtado, J, Lopez-Mendez, B, Stella, S, Montoya, G. | | Deposit date: | 2021-09-16 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis of cyclic oligoadenylate degradation by ancillary Type III CRISPR-Cas ring nucleases.
Nucleic Acids Res., 49, 2021
|
|
7P6M
 
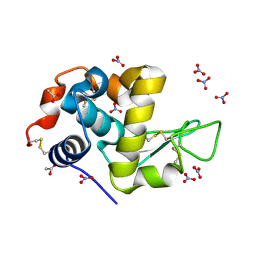 | | Hydrogenated refolded hen egg-white lysozyme | | Descriptor: | ACETATE ION, Lysozyme C, NITRATE ION | | Authors: | Ramos, J, Laux, V, Haertlein, M, Forsyth, V.T, Mossou, E, Larsen, S, Langkilde, A.E. | | Deposit date: | 2021-07-16 | | Release date: | 2021-12-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | The impact of folding modes and deuteration on the atomic resolution structure of hen egg-white lysozyme.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
8RJL
 
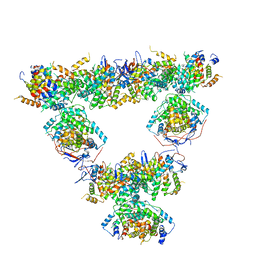 | | Structure of a first order Sierpinski triangle formed by the H369R mutant of the citrate synthase from Synechococcus elongatus | | Descriptor: | Citrate synthase | | Authors: | Lo, Y.K, Bohn, S, Sendker, F.L, Schuller, J.M, Hochberg, G. | | Deposit date: | 2023-12-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Emergence of fractal geometries in the evolution of a metabolic enzyme.
Nature, 628, 2024
|
|
8RJK
 
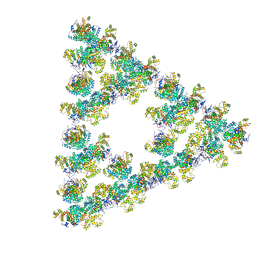 | | Pseudoatomic model of a second-order Sierpinski triangle formed by the citrate synthase from Synechococcus elongatus | | Descriptor: | Citrate synthase | | Authors: | Lo, Y.K, Bohn, S, Sendker, F.L, Schuller, J.M, Hochberg, G. | | Deposit date: | 2023-12-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.91 Å) | | Cite: | Emergence of fractal geometries in the evolution of a metabolic enzyme.
Nature, 628, 2024
|
|
8S5N
 
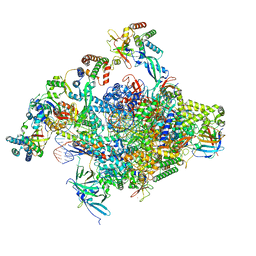 | | RNA polymerase II core initially transcribing complex with an ordered RNA of 12 nt | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-24 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8S55
 
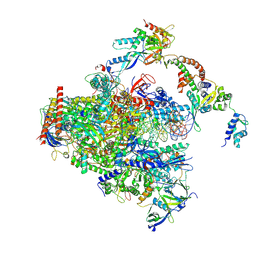 | | RNA polymerase II early elongation complex bound to TFIIE and TFIIF - state a (composite structure) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8RHZ
 
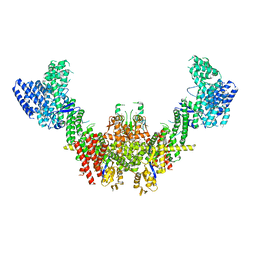 | | Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated conformation - symmetry expanded unneddylated dimer | | Descriptor: | Cullin-9, E3 ubiquitin-protein ligase RBX1, ZINC ION | | Authors: | Hopf, L.V.M, Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2023-12-17 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Noncanonical assembly, neddylation and chimeric cullin-RING/RBR ubiquitylation by the 1.8 MDa CUL9 E3 ligase complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
