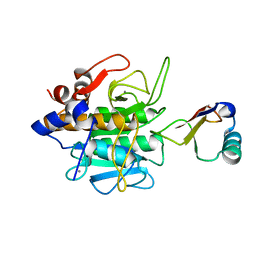
1CSE
 
 | |
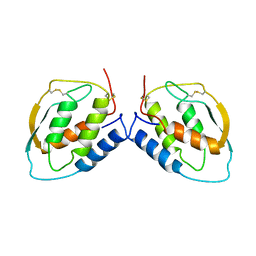
1CSG
 
 | |
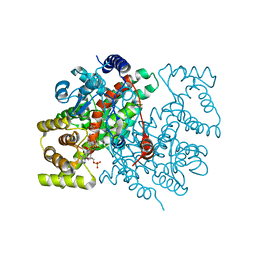
1CSH
 
 | |
1CSI
 
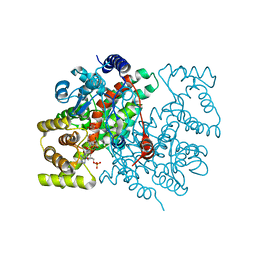 | |
1CSJ
 
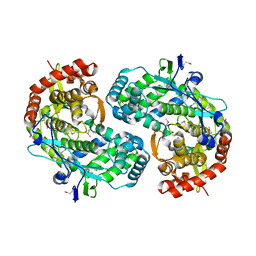 | | CRYSTAL STRUCTURE OF THE RNA-DEPENDENT RNA POLYMERASE OF HEPATITIS C VIRUS | | Descriptor: | HEPATITIS C VIRUS RNA POLYMERASE (NS5B) | | Authors: | Bressanelli, S, Tomei, L, Roussel, A, Incitti, I, Vitale, R.L, Mathieu, M, De Francesco, R, Rey, F.A. | | Deposit date: | 1999-08-18 | | Release date: | 1999-11-08 | | Last modified: | 2013-02-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1CSK
 
 | |
1CSL
 
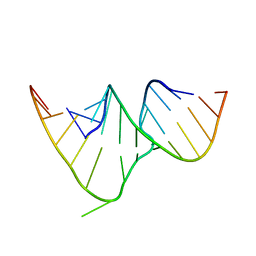 | | CRYSTAL STRUCTURE OF THE RRE HIGH AFFINITY SITE | | Descriptor: | 5'-R(*AP*AP*CP*GP*GP*GP*CP*GP*CP*AP*GP*AP*A)-3', 5'-R(*UP*CP*UP*GP*AP*CP*GP*GP*UP*AP*CP*GP*UP*UP*U)-3' | | Authors: | Ippolito, J.A, Steitz, T.A. | | Deposit date: | 1999-08-18 | | Release date: | 2000-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of the HIV-1 RRE high affinity rev binding site at 1.6 A resolution.
J.Mol.Biol., 295, 2000
|
|
1CSM
 
 | |
1CSN
 
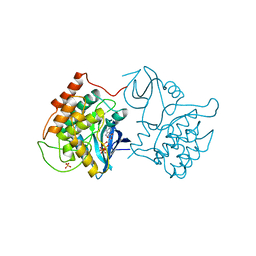 | | BINARY COMPLEX OF CASEIN KINASE-1 WITH MGATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CASEIN KINASE-1, MAGNESIUM ION, ... | | Authors: | Xu, R.-M, Cheng, X. | | Deposit date: | 1995-04-25 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of casein kinase-1, a phosphate-directed protein kinase.
EMBO J., 14, 1995
|
|
1CSO
 
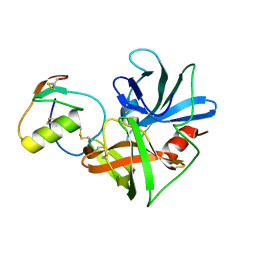 | | CRYSTAL STRUCTURE OF THE OMTKY3 P1 VARIANT OMTKY3-ILE18I IN COMPLEX WITH SGPB | | Descriptor: | OVOMUCOID INHIBITOR, PROTEINASE B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-08-18 | | Release date: | 2000-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Deleterious effects of beta-branched residues in the S1 specificity pocket of Streptomyces griseus proteinase B (SGPB): crystal structures of the turkey ovomucoid third domain variants Ile18I, Val18I, Thr18I, and Ser18I in complex with SGPB.
Protein Sci., 9, 2000
|
|
1CSP
 
 | |
1CSQ
 
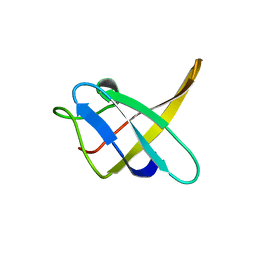 | |
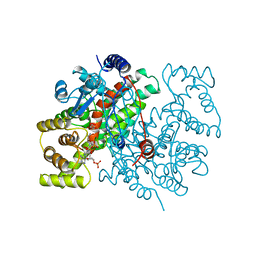
1CSR
 
 | |
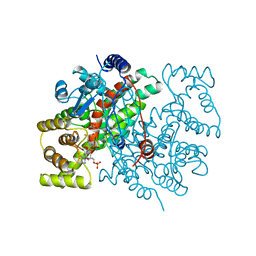
1CSS
 
 | |
1CSU
 
 | |
1CSV
 
 | |
1CSW
 
 | |
1CSX
 
 | |
1CSY
 
 | | SYK TYROSINE KINASE C-TERMINAL SH2 DOMAIN COMPLEXED WITH A PHOSPHOPEPTIDEFROM THE GAMMA CHAIN OF THE HIGH AFFINITY IMMUNOGLOBIN G RECEPTOR, NMR | | Descriptor: | ACETYL-THR-PTR-GLU-THR-LEU-NH2, SYK PROTEIN TYROSINE KINASE | | Authors: | Narula, S.S, Yuan, R.W, Adams, S.E, Green, O.M, Green, J, Phillips, T.B, Zydowsky, L.D, Botfield, M.C, Hatada, M.H, Laird, E.R, Zoller, M.J, Karas, J.L, Dalgarno, D.C. | | Deposit date: | 1995-10-03 | | Release date: | 1996-11-08 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal SH2 domain of the human tyrosine kinase Syk complexed with a phosphotyrosine pentapeptide.
Structure, 3, 1995
|
|
1CSZ
 
 | | SYK TYROSINE KINASE C-TERMINAL SH2 DOMAIN COMPLEXED WITH A PHOSPHOPEPTIDEFROM THE GAMMA CHAIN OF THE HIGH AFFINITY IMMUNOGLOBIN G RECEPTOR, NMR | | Descriptor: | ACETYL-THR-PTR-GLU-THR-LEU-NH2, SYK PROTEIN TYROSINE KINASE | | Authors: | Narula, S.S, Yuan, R.W, Adams, S.E, Green, O.M, Green, J, Phillips, T.B, Zydowsky, L.D, Botfield, M.C, Hatada, M.H, Laird, E.R, Zoller, M.J, Karas, J.L, Dalgarno, D.C. | | Deposit date: | 1995-10-03 | | Release date: | 1996-11-08 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal SH2 domain of the human tyrosine kinase Syk complexed with a phosphotyrosine pentapeptide.
Structure, 3, 1995
|
|
1CT0
 
 | | CRYSTAL STRUCTURE OF THE OMTKY3 P1 VARIANT OMTKY3-SER18I IN COMPLEX WITH SGPB | | Descriptor: | OVOMUCOID INHIBITOR, PROTEINASE B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-08-18 | | Release date: | 2000-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Deleterious effects of beta-branched residues in the S1 specificity pocket of Streptomyces griseus proteinase B (SGPB): crystal structures of the turkey ovomucoid third domain variants Ile18I, Val18I, Thr18I, and Ser18I in complex with SGPB.
Protein Sci., 9, 2000
|
|
1CT1
 
 | | CHOLERA TOXIN B-PENTAMER MUTANT G33R BOUND TO RECEPTOR PENTASACCHARIDE | | Descriptor: | CHLORIDE ION, CHOLERA TOXIN, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Merritt, E.A, Hol, W.G.J. | | Deposit date: | 1997-06-03 | | Release date: | 1997-10-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural studies of receptor binding by cholera toxin mutants.
Protein Sci., 6, 1997
|
|
1CT2
 
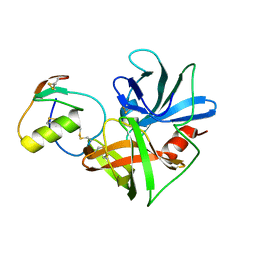 | | CRYSTAL STRUCTURE OF THE OMTKY3 P1 VARIANT OMTKY3-THR18I IN COMPLEX WITH SGPB | | Descriptor: | OVOMUCOID INHIBITOR, PROTEINASE B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N. | | Deposit date: | 1999-08-18 | | Release date: | 2000-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Deleterious effects of beta-branched residues in the S1 specificity pocket of Streptomyces griseus proteinase B (SGPB): crystal structures of the turkey ovomucoid third domain variants Ile18I, Val18I, Thr18I, and Ser18I in complex with SGPB.
Protein Sci., 9, 2000
|
|
1CT4
 
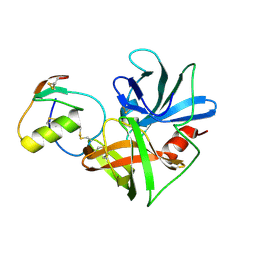 | | CRYSTAL STRUCTURE OF THE OMTKY3 P1 VARIANT OMTKY3-VAL18I IN COMPLEX WITH SGPB | | Descriptor: | OVOMUCOID INHIBITOR, PROTEINASE B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-08-18 | | Release date: | 2000-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Deleterious effects of beta-branched residues in the S1 specificity pocket of Streptomyces griseus proteinase B (SGPB): crystal structures of the turkey ovomucoid third domain variants Ile18I, Val18I, Thr18I, and Ser18I in complex with SGPB.
Protein Sci., 9, 2000
|
|
1CT5
 
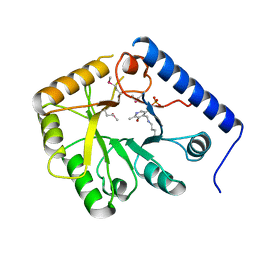 | | CRYSTAL STRUCTURE OF YEAST HYPOTHETICAL PROTEIN YBL036C-SELENOMET CRYSTAL | | Descriptor: | PROTEIN (YEAST HYPOTHETICAL PROTEIN, SELENOMET), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Eswaramoorthy, S, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 1999-08-18 | | Release date: | 1999-09-02 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a yeast hypothetical protein selected by a structural genomics approach.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
