3JSY
 
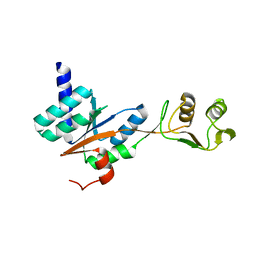 | | N-terminal fragment of ribosomal protein L10 from Methanococcus jannaschii | | Descriptor: | Acidic ribosomal protein P0 homolog | | Authors: | Kravchenko, O, Mitroshin, I, Nikulin, A.D, Piendl, W, Garber, M, Nikonov, S. | | Deposit date: | 2009-09-11 | | Release date: | 2010-05-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a two-domain N-terminal fragment of ribosomal protein L10 from Methanococcus jannaschii reveals a specific piece of the archaeal ribosomal stalk
J.Mol.Biol., 399, 2010
|
|
1U9Y
 
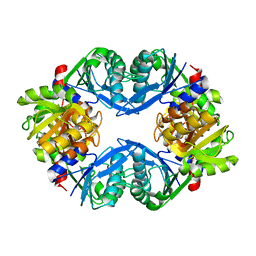 | | Crystal Structure of Phosphoribosyl Diphosphate Synthase from Methanocaldococcus jannaschii | | Descriptor: | Ribose-phosphate pyrophosphokinase | | Authors: | Kadziola, A, Johansson, E, Jepsen, C.H, McGuire, J, Larsen, S, Hove-Jensen, B. | | Deposit date: | 2004-08-11 | | Release date: | 2005-08-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Novel class III phosphoribosyl diphosphate synthase: structure and properties of the tetrameric, phosphate-activated, non-allosterically inhibited enzyme from Methanocaldococcus jannaschii
J.Mol.Biol., 354, 2005
|
|
9IMU
 
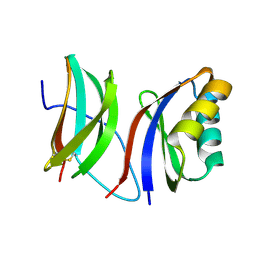 | |
1AM9
 
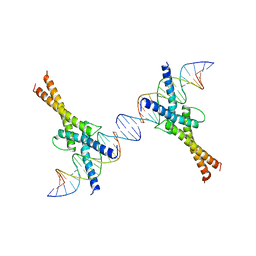 | | HUMAN SREBP-1A BOUND TO LDL RECEPTOR PROMOTER | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*AP*GP*AP*TP*CP*AP*CP*CP*CP*CP*AP*CP*T P*GP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*CP*AP*GP*TP*GP*GP*GP*GP*TP*GP*AP*TP*CP*T )-3'), MAGNESIUM ION, ... | | Authors: | Parraga, A, Burley, S.K. | | Deposit date: | 1997-06-25 | | Release date: | 1998-07-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Co-crystal structure of sterol regulatory element binding protein 1a at 2.3 A resolution.
Structure, 6, 1998
|
|
6T1Y
 
 | | Cryo-EM structure of phalloidin-stabilized F-actin (copolymerized) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Merino, F, Raunser, S. | | Deposit date: | 2019-10-07 | | Release date: | 2020-03-04 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Effects and Functional Implications of Phalloidin and Jasplakinolide Binding to Actin Filaments.
Structure, 28, 2020
|
|
6TLK
 
 | | Structure of methylene-tetrahydromethanopterin dehydrogenase from Methylorubrum extorquens AM1 in an open conformation containing NADP+ and methylene-H4MPT | | Descriptor: | 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, Bifunctional protein MdtA, CHLORIDE ION, ... | | Authors: | Wagner, T, Huang, G, Demmer, U, Warkentin, E, Ermler, U, Shima, S. | | Deposit date: | 2019-12-02 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Hydride Transfer Process in NADP-dependent Methylene-tetrahydromethanopterin Dehydrogenase.
J.Mol.Biol., 432, 2020
|
|
6TM3
 
 | | Structure of methylene-tetrahydromethanopterin dehydrogenase from Methylorubrum extorquens AM1 in a close conformation containing NADP+ and methylene-H4MPT | | Descriptor: | 1,2-ETHANEDIOL, 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, Bifunctional protein MdtA, ... | | Authors: | Wagner, T, Huang, G, Demmer, U, Warkentin, E, Ermler, U, Shima, S. | | Deposit date: | 2019-12-03 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | The Hydride Transfer Process in NADP-dependent Methylene-tetrahydromethanopterin Dehydrogenase.
J.Mol.Biol., 432, 2020
|
|
6T25
 
 | | Cryo-EM structure of phalloidin-Alexa Flour-546-stabilized F-actin (copolymerized) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Merino, F, Raunser, S. | | Deposit date: | 2019-10-07 | | Release date: | 2020-03-04 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Effects and Functional Implications of Phalloidin and Jasplakinolide Binding to Actin Filaments.
Structure, 28, 2020
|
|
6T23
 
 | | Cryo-EM structure of jasplakinolide-stabilized F-actin (aged) | | Descriptor: | (4~{R},7~{R},10~{S},13~{S},15~{E},19~{S})-10-(4-azanylbutyl)-4-(4-hydroxyphenyl)-7-(1~{H}-indol-3-ylmethyl)-8,13,15,19-tetramethyl-1-oxa-5,8,11-triazacyclononadec-15-ene-2,6,9,12-tetrone, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Pospich, S, Merino, F, Raunser, S. | | Deposit date: | 2019-10-07 | | Release date: | 2020-03-04 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Effects and Functional Implications of Phalloidin and Jasplakinolide Binding to Actin Filaments.
Structure, 28, 2020
|
|
6TTU
 
 | | Ubiquitin Ligation to substrate by a cullin-RING E3 ligase at 3.7A resolution: NEDD8-CUL1-RBX1 N98R-SKP1-monomeric b-TRCP1dD-IkBa-UB~UBE2D2 | | Descriptor: | CYS-LYS-LYS-ALA-ARG-HIS-ASP-SEP-GLY, Cullin-1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2019-12-30 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | NEDD8 nucleates a multivalent cullin-RING-UBE2D ubiquitin ligation assembly.
Nature, 578, 2020
|
|
6YKB
 
 | | [Fe]-hydrogenase from Methanolacinia paynteri in complex with GMP at 1.55-A resolution | | Descriptor: | 5,10-methenyltetrahydromethanopterin [Fe]-hydrogenase, FORMIC ACID, GLYCEROL, ... | | Authors: | Wagner, T, Huang, G, Arriaza-Gallardo, F.J, Shima, S. | | Deposit date: | 2020-04-06 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Hydride Transfer Process in NADP-dependent Methylene-tetrahydromethanopterin Dehydrogenase.
J.Mol.Biol., 432, 2020
|
|
6YKA
 
 | | Asymmetric [Fe]-hydrogenase from Methanolacinia paynteri apo and in complex with FeGP at 2.1-A resolution | | Descriptor: | 1,2-ETHANEDIOL, 5,10-methenyltetrahydromethanopterin hydrogenase, GLYCEROL, ... | | Authors: | Wagner, T, Huang, G, Arriaza-Gallardo, F.J, Shima, S. | | Deposit date: | 2020-04-06 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Hydride Transfer Process in NADP-dependent Methylene-tetrahydromethanopterin Dehydrogenase.
J.Mol.Biol., 432, 2020
|
|
8AXS
 
 | | Sialidases and Fucosidases of Akkermansia muciniphila are key for rapid growth on colonic mucin and nutrient sharing amongst mucin-associated human gut microbiota | | Descriptor: | 1,2-ETHANEDIOL, 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CALCIUM ION, ... | | Authors: | Sakanaka, H, Nielsen, T.S, Pichler, M.J, Nordberg Karlsson, E, Abou Hachem, M, Morth, J.P. | | Deposit date: | 2022-08-31 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Sialidases and fucosidases of Akkermansia muciniphila are crucial for growth on mucin and nutrient sharing with mucus-associated gut bacteria.
Nat Commun, 14, 2023
|
|
8AXI
 
 | | Sialidases and Fucosidases of Akkermansia muciniphila are key for rapid growth on colonic mucin and nutrient sharing amongst mucin-associated human gut microbiota | | Descriptor: | 1,2-ETHANEDIOL, 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CALCIUM ION, ... | | Authors: | Sakanaka, H, Nielsen, T.S, Pichler, M.J, Nordberg Karlsson, E, Abou Hachem, M, Morth, J.P. | | Deposit date: | 2022-08-31 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Sialidases and fucosidases of Akkermansia muciniphila are crucial for growth on mucin and nutrient sharing with mucus-associated gut bacteria.
Nat Commun, 14, 2023
|
|
8AWI
 
 | | Crystal structure of Human Transthyretin at 1.15 Angstrom resolution | | Descriptor: | SODIUM ION, Transthyretin | | Authors: | Derbyshire, D.J, Hammarstrom, P, von Castelmur, E, Begum, A. | | Deposit date: | 2022-08-29 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Transthyretin Binding Mode Dichotomy of Fluorescent trans -Stilbene Ligands.
Acs Chem Neurosci, 14, 2023
|
|
6YK9
 
 | | [Fe]-hydrogenase from Methanolacinia paynteri with bound guanylylpyridinol at 1.7-A resolution | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-{[2-(carboxymethyl)-6-hydroxy-3,5-dimethylpyridin-4-yl]oxy}(hydroxy)phosphoryl]guanosine, 5,10-methenyltetrahydromethanopterin hydrogenase, ... | | Authors: | Wagner, T, Huang, G, Arriaza-Gallardo, F.J, Shima, S. | | Deposit date: | 2020-04-06 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Hydride Transfer Process in NADP-dependent Methylene-tetrahydromethanopterin Dehydrogenase.
J.Mol.Biol., 432, 2020
|
|
8PF9
 
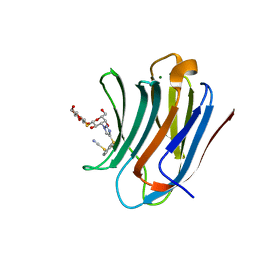 | | Galectin-3C in complex with a triazolesulfane derivative | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-[4-[oxidanyl(phenyl)-$l^{3}-sulfanyl]-1,2,3-triazol-1-yl]oxan-2-yl]sulfanyl-oxane-3,4,5-triol, Galectin-3, MAGNESIUM ION, ... | | Authors: | Kumar, R, Mahanti, M, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2023-06-15 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Ligand Sulfur Oxidation State Progressively Alters Galectin-3-Ligand Complex Conformations To Induce Affinity-Influencing Hydrogen Bonds.
J.Med.Chem., 66, 2023
|
|
8PBF
 
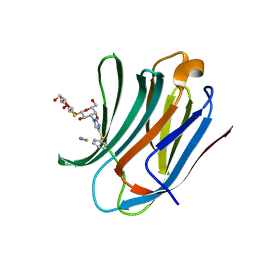 | | Galectin-3C in complex with a triazolesulfane derivative | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-(4-phenylsulfanyl-1,2,3-triazol-1-yl)oxan-2-yl]sulfanyl-oxane-3,4,5-triol, Galectin-3, THIOCYANATE ION | | Authors: | Kumar, R, Mahanti, M. | | Deposit date: | 2023-06-09 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Ligand Sulfur Oxidation State Progressively Alters Galectin-3-Ligand Complex Conformations To Induce Affinity-Influencing Hydrogen Bonds.
J.Med.Chem., 66, 2023
|
|
8PFF
 
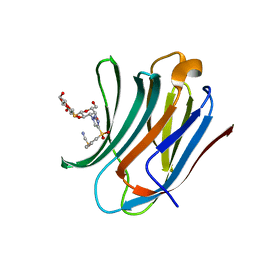 | | Galectin-3C in complex with a triazolesulfone derivative | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-[4-(phenylsulfonyl)-1,2,3-triazol-1-yl]oxan-2-yl]sulfanyl-oxane-3,4,5-triol, Galectin-3, MAGNESIUM ION, ... | | Authors: | Kumar, R, Mahanti, M, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2023-06-15 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Ligand Sulfur Oxidation State Progressively Alters Galectin-3-Ligand Complex Conformations To Induce Affinity-Influencing Hydrogen Bonds.
J.Med.Chem., 66, 2023
|
|
8B8E
 
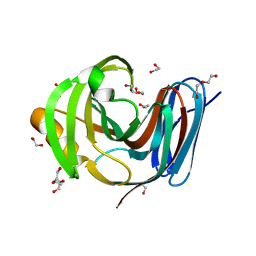 | | Wild-type GH11 from Blastobotrys mokoenaii | | Descriptor: | 1,2-ETHANEDIOL, BmGH11, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Coleman, T, Ravn, J.L, Larsbrink, J. | | Deposit date: | 2022-10-04 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Yeasts Have Evolved Divergent Enzyme Strategies To Deconstruct and Metabolize Xylan.
Microbiol Spectr, 11, 2023
|
|
6ZF8
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 1-[3-[2-hydroxy-2-oxoethyl-(3-methoxyphenyl)sulfonyl-amino]phenyl]-5-[(1~{S},2~{S})-2-phenylcyclopropyl]pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
8B48
 
 | | Structure of Lentithecium fluviatile carbohydrate esterase from the CE15 family (LfCE15C) | | Descriptor: | Carbohydrate esterase family 15 protein, FORMIC ACID, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scholzen, K, Mazurkewich, S, Poulsen, J.C.N, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2022-09-20 | | Release date: | 2023-06-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and functional investigation of a fungal member of carbohydrate esterase family 15 with potential specificity for rare xylans.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8AVI
 
 | | Crystal structure of IsdG from Bacillus cereus in complex with heme | | Descriptor: | 1,3-PROPANDIOL, 1,4-BUTANEDIOL, Heme-degrading monooxygenase, ... | | Authors: | Andersen, H.K, Hammerstad, M, Hersleth, H.-P. | | Deposit date: | 2022-08-26 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional Diversity of Homologous Oxidoreductases-Tuning of Substrate Specificity by a FAD-Stacking Residue for Iron Acquisition and Flavodoxin Reduction.
Antioxidants, 12, 2023
|
|
6YU9
 
 | |
8AVH
 
 | | Crystal structure of IsdG from Bacillus cereus | | Descriptor: | 1,3-PROPANDIOL, Heme-degrading monooxygenase | | Authors: | Andersen, H.K, Hammerstad, M, Hersleth, H.-P. | | Deposit date: | 2022-08-26 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional Diversity of Homologous Oxidoreductases-Tuning of Substrate Specificity by a FAD-Stacking Residue for Iron Acquisition and Flavodoxin Reduction.
Antioxidants, 12, 2023
|
|
