5WO0
 
 | | DNA polymerase beta substrate complex with incoming 5-FodUTP | | Descriptor: | 2'-deoxy-5-formyluridine 5'-(tetrahydrogen triphosphate), CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schaich, M.A, Smith, M.R, Cloud, A.S, Holloran, S.M, Freudenthal, B.D. | | Deposit date: | 2017-08-01 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of a DNA Polymerase Inserting Therapeutic Nucleotide Analogues.
Chem. Res. Toxicol., 30, 2017
|
|
5EE7
 
 | | Crystal structure of the human glucagon receptor (GCGR) in complex with the antagonist MK-0893 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 3-[[4-[(1~{S})-1-[3-[3,5-bis(chloranyl)phenyl]-5-(6-methoxynaphthalen-2-yl)pyrazol-1-yl]ethyl]phenyl]carbonylamino]propanoic acid, Glucagon receptor,Endolysin,Glucagon receptor, ... | | Authors: | Jazayeri, A, Dore, A.S, Lamb, D, Krishnamurthy, H, Southall, S.M, Baig, A.H, Bortolato, A, Koglin, M, Robertson, N.J, Errey, J.C, Andrews, S.P, Brown, A.J.H, Cooke, R.M, Weir, M, Marshall, F.H. | | Deposit date: | 2015-10-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Extra-helical binding site of a glucagon receptor antagonist.
Nature, 533, 2016
|
|
5LIX
 
 | | Crystal structure of human AKR1B10 complexed with NADP+ and the inhibitor MK184 | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Fanfrlik, J, Kamlar, M, Vesely, J, Hobza, P, Podjarny, A. | | Deposit date: | 2016-07-15 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | IDD388 Polyhalogenated Derivatives as Probes for an Improved Structure-Based Selectivity of AKR1B10 Inhibitors.
Acs Chem.Biol., 11, 2016
|
|
6O0H
 
 | |
4LQR
 
 | | Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens | | Descriptor: | CALCIUM ION, Glycosyl hydrolase, family 31/fibronectin type III domain protein | | Authors: | Grondin, J.M, Furness, H.S, Duan, D, Spencer, C.A, Allingham, J.S, Smith, S.P. | | Deposit date: | 2013-07-19 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
Plos One, 12, 2017
|
|
5W3R
 
 | | SH2B1 SH2 Domain | | Descriptor: | PHENOL, PHOSPHATE ION, SH2B adapter protein 1 | | Authors: | McKercher, M.A, Wuttke, D.S. | | Deposit date: | 2017-06-08 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.386 Å) | | Cite: | Diversity in peptide recognition by the SH2 domain of SH2B1.
Proteins, 86, 2018
|
|
5LIU
 
 | | Crystal structure of human AKR1B10 complexed with NADP+ and the inhibitor IDD388 | | Descriptor: | (2-{[(4-BROMO-2-FLUOROBENZYL)AMINO]CARBONYL}-5-CHLOROPHENOXY)ACETIC ACID, 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, ... | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Fanfrlik, J, Kamlar, M, Vesely, J, Hobza, P, Podjarny, A. | | Deposit date: | 2016-07-15 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | IDD388 Polyhalogenated Derivatives as Probes for an Improved Structure-Based Selectivity of AKR1B10 Inhibitors.
Acs Chem.Biol., 11, 2016
|
|
4MDK
 
 | | Cdc34-ubiquitin-CC0651 complex | | Descriptor: | 4,5-dideoxy-5-(3',5'-dichlorobiphenyl-4-yl)-4-[(methoxyacetyl)amino]-L-arabinonic acid, Ubiquitin, Ubiquitin-conjugating enzyme E2 R1 | | Authors: | Ceccarelli, D.F, Orlicky, S, Tyers, M, Sicheri, F. | | Deposit date: | 2013-08-22 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6095 Å) | | Cite: | E2 enzyme inhibition by stabilization of a low-affinity interface with ubiquitin.
Nat.Chem.Biol., 10, 2014
|
|
5WLL
 
 | |
5LMD
 
 | | The crystal structure of hCA II in complex with a benzoxaborole inhibitor | | Descriptor: | 1-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-4-yl]-3-(2-methoxy-5-methyl-phenyl)urea, Carbonic anhydrase 2, ZINC ION | | Authors: | De Simone, G, Alterio, V, Esposito, D, Di Fiore, A. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Benzoxaborole as a new chemotype for carbonic anhydrase inhibition.
Chem.Commun.(Camb.), 52, 2016
|
|
6TJQ
 
 | | Crystal Structure of Recombinant GBA in Complex with 2-Deoxy-2-fluoro-beta-D-glucopyranoside | | Descriptor: | (2~{R},3~{S},4~{S},5~{S})-5-fluoranyl-2-(hydroxymethyl)oxane-3,4-diol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2019-11-26 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A baculoviral system for the production of human beta-glucocerebrosidase enables atomic resolution analysis.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
9BBH
 
 | | Co-crystal structure of human DDB1 bound to fragment UB028670 | | Descriptor: | 1,2-ETHANEDIOL, 4-methoxy-1H-indole, DNA damage-binding protein 1, ... | | Authors: | Zeng, H, Dong, A, Frommlet, A, Seitova, A, Loppnau, P, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-05 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Co-crystal structure of human DDB1 bound to fragment UB028670
To be published
|
|
4JGR
 
 | | The crystal structure of sporulation kinase D mutant sensor domain, R131A, from Bacillus subtilis subsp at 2.4A resolution | | Descriptor: | ACETIC ACID, GLYCEROL, Sporulation kinase D | | Authors: | Wu, R, Schiffer, M, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-03-01 | | Release date: | 2013-05-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insight into the sporulation phosphorelay: Crystal structure of the sensor domain of Bacillus subtilis histidine kinase, KinD.
Protein Sci., 22, 2013
|
|
5TGA
 
 | |
7JQL
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Bac7-001, mRNA, and deacylated P-site tRNA at 3.00A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mardirossian, M, Sola, R, Beckert, B, Valencic, E, Collis, D.W.P, Borisek, J, Armas, F, Di Stasi, A, Buchmann, J, Syroegin, E.A, Polikanov, Y.S, Magistrato, A, Hilpert, K, Wilson, D.N, Scocchi, M. | | Deposit date: | 2020-08-11 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Peptide Inhibitors of Bacterial Protein Synthesis with Broad Spectrum and SbmA-Independent Bactericidal Activity against Clinical Pathogens.
J.Med.Chem., 63, 2020
|
|
4JGP
 
 | | The crystal structure of sporulation kinase D sensor domain from Bacillus subtilis subsp in complex with pyruvate at 2.0A resolution | | Descriptor: | PYRUVIC ACID, Sporulation kinase D | | Authors: | Wu, R, Schiffer, M, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-03-01 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Insight into the sporulation phosphorelay: Crystal structure of the sensor domain of Bacillus subtilis histidine kinase, KinD.
Protein Sci., 22, 2013
|
|
4JGQ
 
 | | The crystal structure of sporulation kinase D mutant sensor domain, r131a, from Bacillus subtilis subsp in co-crystallization with pyruvate | | Descriptor: | ACETIC ACID, Sporulation kinase D | | Authors: | Wu, R, Schiffer, M, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-03-01 | | Release date: | 2013-05-15 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Insight into the sporulation phosphorelay: Crystal structure of the sensor domain of Bacillus subtilis histidine kinase, KinD.
Protein Sci., 22, 2013
|
|
7JQM
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Bac7-002, mRNA, and deacylated P-site tRNA at 3.05A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mardirossian, M, Sola, R, Beckert, B, Valencic, E, Collis, D.W.P, Borisek, J, Armas, F, Di Stasi, A, Buchmann, J, Syroegin, E.A, Polikanov, Y.S, Magistrato, A, Hilpert, K, Wilson, D.N, Scocchi, M. | | Deposit date: | 2020-08-11 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Peptide Inhibitors of Bacterial Protein Synthesis with Broad Spectrum and SbmA-Independent Bactericidal Activity against Clinical Pathogens.
J.Med.Chem., 63, 2020
|
|
4JGO
 
 | | The crystal structure of sporulation kinase d sensor domain from Bacillus subtilis subsp. | | Descriptor: | GLYCEROL, PYRUVIC ACID, Sporulation kinase D | | Authors: | Wu, R, Schiffer, M, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-03-01 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Insight into the sporulation phosphorelay: Crystal structure of the sensor domain of Bacillus subtilis histidine kinase, KinD.
Protein Sci., 22, 2013
|
|
5V8I
 
 | | Thermus thermophilus 70S ribosome lacking ribosomal protein uS17 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Gregory, S.T, Jogl, G. | | Deposit date: | 2017-03-22 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural robustness of the ribosome inferred from X-ray crystal structures of the 30S ribosomal subunit and the 70S ribosome lacking ribosomal protein uS17
To be published
|
|
5VP2
 
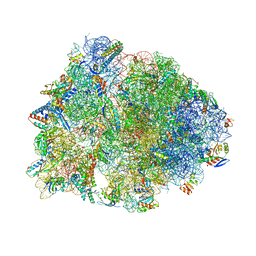 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with madumycin II and bound to mRNA and A-, P- and E-site tRNAs at 2.8A resolution | | Descriptor: | 16S ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Osterman, I.A, Khabibullina, N.F, Komarova, E.S, Kasatsky, P, Kartsev, V.G, Bogdanov, A.A, Dontsova, O.A, Konevega, A.L, Sergiev, P.V, Polikanov, Y.S. | | Deposit date: | 2017-05-04 | | Release date: | 2017-06-28 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Madumycin II inhibits peptide bond formation by forcing the peptidyl transferase center into an inactive state.
Nucleic Acids Res., 45, 2017
|
|
3N4A
 
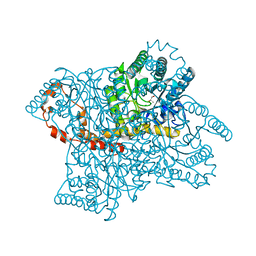 | | Crystal structure of D-Xylose Isomerase in complex with S-1,2-Propandiol | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, S-1,2-PROPANEDIOL, ... | | Authors: | Behnen, J, Heine, A, Klebe, G. | | Deposit date: | 2010-05-21 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Experimental and computational active site mapping as a starting point to fragment-based lead discovery.
Chemmedchem, 7, 2012
|
|
3E85
 
 | | Crystal Structure of Pathogenesis-related Protein LlPR-10.2B from yellow lupine in complex with Diphenylurea | | Descriptor: | 1,3-DIPHENYLUREA, PR10.2B, SODIUM ION | | Authors: | Fernandes, H.C, Bujacz, G, Bujacz, A, Sikorski, M.M, Jaskolski, M. | | Deposit date: | 2008-08-19 | | Release date: | 2009-03-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cytokinin-induced structural adaptability of a Lupinus luteus PR-10 protein.
Febs J., 276, 2009
|
|
4WCX
 
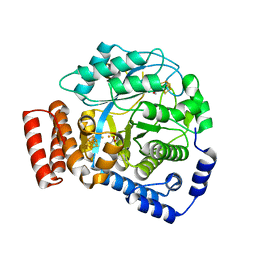 | | Crystal structure of HydG: A maturase of the [FeFe]-hydrogenase | | Descriptor: | ALANINE, Biotin and thiamin synthesis associated, FE (III) ION, ... | | Authors: | Dinis, P.C, Harmer, J.E, Driesener, R.C, Roach, P.L. | | Deposit date: | 2014-09-05 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | X-ray crystallographic and EPR spectroscopic analysis of HydG, a maturase in [FeFe]-hydrogenase H-cluster assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3EOB
 
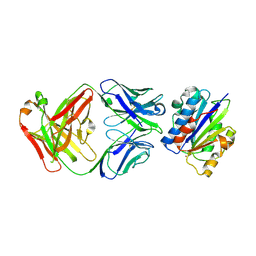 | |
