7SAR
 
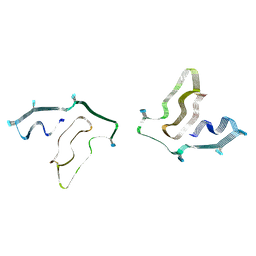 | | Cryo-EM structure of TMEM106B fibrils extracted from a FTLD-TDP patient, polymorph 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Cao, Q, Jiang, Y, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-09-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Amyloid fibrils in FTLD-TDP are composed of TMEM106B and not TDP-43.
Nature, 605, 2022
|
|
7LD9
 
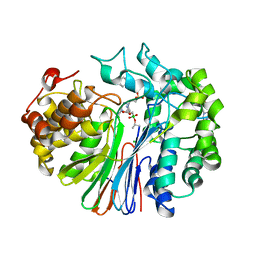 | | Structure of human GGT1 in complex with ABBA | | Descriptor: | (2R)-2-amino-4-boronobutanoic acid, (2S)-2-amino-4-boronobutanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Terzyan, S.S, Hanigan, M. | | Deposit date: | 2021-01-12 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Design and evaluation of novel analogs of 2-amino-4-boronobutanoic acid (ABBA) as inhibitors of human gamma-glutamyl transpeptidase.
Bioorg.Med.Chem., 73, 2022
|
|
7UHW
 
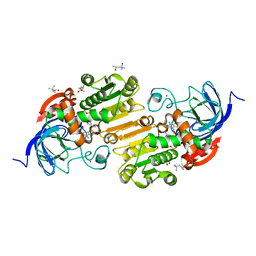 | | Horse liver alcohol dehydrogenase G173A, complexed with NAD+ and 2,3,4,5,6-pentafluorobenzyl alcohol at 120 K | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2,3,4,5,6-PENTAFLUOROBENZYL ALCOHOL, Alcohol dehydrogenase E chain, ... | | Authors: | Plapp, B.V, Gakhar, L. | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Dependence of crystallographic atomic displacement factors on temperature (25-150 K) for complexes of horse liver alcohol dehydrogenases
Acta Crystallogr.,Sect.D, D78, 2022
|
|
3AKD
 
 | |
3K3K
 
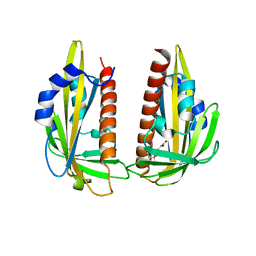 | | Crystal structure of dimeric abscisic acid (ABA) receptor pyrabactin resistance 1 (PYR1) with ABA-bound closed-lid and ABA-free open-lid subunits | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYR1 | | Authors: | Arvai, A.S, Hitomi, K, Getzoff, E.D. | | Deposit date: | 2009-10-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural mechanism of abscisic acid binding and signaling by dimeric PYR1.
Science, 326, 2009
|
|
7LRX
 
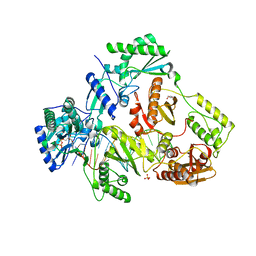 | | Structure of HIV-1 Reverse Transcriptase in complex with DNA, L-dCTP, and CA(2+) ion | | Descriptor: | 4-amino-1-{2-deoxy-5-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-L-erythro-pentofuranosyl}pyrimidin-2(1H)-one, AMMONIUM ION, CALCIUM ION, ... | | Authors: | Hoang, A, Ruiz, F.X, Arnold, E. | | Deposit date: | 2021-02-17 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of HIV inhibition by L-nucleosides: Opportunities for drug development and repurposing.
Drug Discov Today, 27, 2022
|
|
7UHX
 
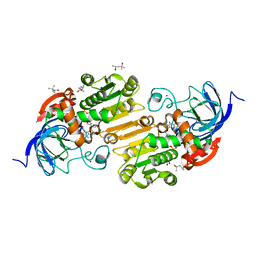 | | Horse liver alcohol dehydrogenase G173A, complexed with NAD+ and 2,3,4,5,6-pentafluorobenzyl alcohol at 150 K | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2,3,4,5,6-PENTAFLUOROBENZYL ALCOHOL, Alcohol dehydrogenase E chain, ... | | Authors: | Plapp, B.V, Gakhar, L. | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Dependence of crystallographic atomic displacement factors on temperature (25-150 K) for complexes of horse liver alcohol dehydrogenases
Acta Crystallogr.,Sect.D, D78, 2022
|
|
7UDD
 
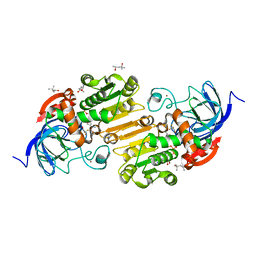 | | Horse liver alcohol dehydrogenase complexed with NAD+ and 2,2,2-trifluoroethanol at 150 K | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Alcohol dehydrogenase E chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), ... | | Authors: | Plapp, B.V, Gakhar, L. | | Deposit date: | 2022-03-18 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Dependence of crystallographic atomic displacement factors on temperature (25-150 K) for complexes of horse liver alcohol dehydrogenases
Acta Crystallogr.,Sect.D, D78, 2022
|
|
7BIK
 
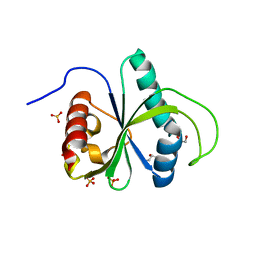 | | Crystal structure of YTHDF2 in complex with m6Am | | Descriptor: | (2~{R},3~{S},4~{R},5~{R})-2-(hydroxymethyl)-4-methoxy-5-[6-(methylamino)purin-9-yl]oxolan-3-ol, GLYCEROL, SULFATE ION, ... | | Authors: | Wang, X, Caflisch, A. | | Deposit date: | 2021-01-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of YTHDF2 in complex with m6Am
To Be Published
|
|
3K4T
 
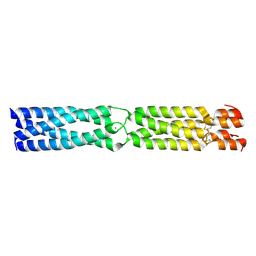 | |
7BCY
 
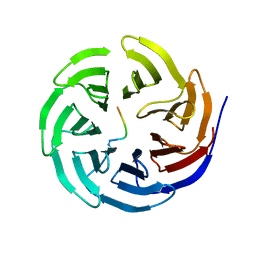 | |
7D0E
 
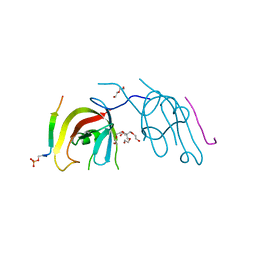 | | Crystal structure of FIP200 Claw/p-CCPG1 FIR2 | | Descriptor: | 3-(2-hydroxyethyloxy)-2-[2-(2-hydroxyethyloxy)ethoxymethyl]-2-(2-hydroxyethyloxymethyl)propan-1-ol, Cell cycle progression protein 1 FIR2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhou, Z.X, Pan, L.F. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphorylation regulates the binding of autophagy receptors to FIP200 Claw domain for selective autophagy initiation.
Nat Commun, 12, 2021
|
|
5MOF
 
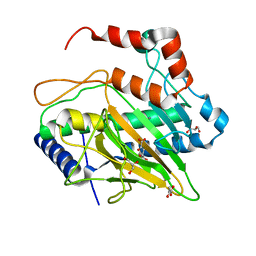 | | Ethylene Forming Enzyme from Pseudomonas syringae pv. phaseolicola - I222 crystal form in complex with manganese and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, CHLORIDE ION, ... | | Authors: | McDonough, M.A, Zhang, Z, Schofield, C.J. | | Deposit date: | 2016-12-14 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and stereoelectronic insights into oxygenase-catalyzed formation of ethylene from 2-oxoglutarate.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7UHV
 
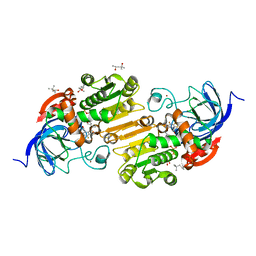 | | Horse liver alcohol dehydrogenase G173A, complexed with NAD+ and 2,3,4,5,6-pentafluorobenzyl alcohol at 50 K | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2,3,4,5,6-PENTAFLUOROBENZYL ALCOHOL, Alcohol dehydrogenase E chain, ... | | Authors: | Plapp, B.V, Gakhar, L. | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Dependence of crystallographic atomic displacement factors on temperature (25-150 K) for complexes of horse liver alcohol dehydrogenases
Acta Crystallogr.,Sect.D, D78, 2022
|
|
7DAD
 
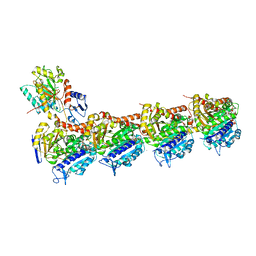 | | EPD in complex with tubulin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, Y, Wu, C. | | Deposit date: | 2020-10-16 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | High-resolution X-ray structure of three microtubule-stabilizing agents in complex with tubulin provide a rationale for drug design.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
2JSH
 
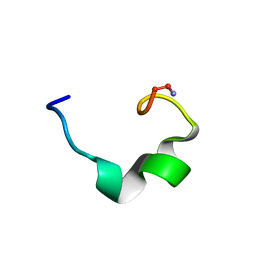 | | obestatin NMR structure in SDS/DPC micellar solution | | Descriptor: | Appetite-regulating hormone, Obestatin | | Authors: | D'Ursi, A.M, Scrima, M, Esposito, C, Campiglia, P. | | Deposit date: | 2007-07-05 | | Release date: | 2008-10-21 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Obestatin conformational features: a strategy to unveil obestatin's biological role?
Biochem.Biophys.Res.Commun., 363, 2007
|
|
3AJP
 
 | | Crystal structure of human H ferritin E140A mutant | | Descriptor: | Ferritin heavy chain, MAGNESIUM ION | | Authors: | Masuda, T, Mikami, B. | | Deposit date: | 2010-06-11 | | Release date: | 2010-08-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | The universal mechanism for iron translocation to the ferroxidase site in ferritin, which is mediated by the well conserved transit site
Biochem.Biophys.Res.Commun., 400, 2010
|
|
7BGT
 
 | | Mason-Pfizer Monkey Virus Protease mutant C7A/D26N/C106A in complex with peptidomimetic inhibitor | | Descriptor: | ACETATE ION, Gag-Pro-Pol polyprotein, PENTAETHYLENE GLYCOL, ... | | Authors: | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2021-01-08 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of inhibitor complexes of M-PMV protease with visible flap loops.
Protein Sci., 30, 2021
|
|
7D4P
 
 | | Structure of human TRPC5 in complex with clemizole | | Descriptor: | (2S)-2-(hexadecanoyloxy)-3-hydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1-[(4-chlorophenyl)methyl]-2-(pyrrolidin-1-ylmethyl)benzimidazole, ... | | Authors: | Chen, L, Song, K, Wei, M, Guo, W. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-31 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for human TRPC5 channel inhibition by two distinct inhibitors.
Elife, 10, 2021
|
|
7CZG
 
 | | Crystal structure of FIP200 Claw domain apo form | | Descriptor: | DI(HYDROXYETHYL)ETHER, RB1-inducible coiled-coil protein 1 | | Authors: | Zhou, Z.X, Pan, L.F. | | Deposit date: | 2020-09-08 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Phosphorylation regulates the binding of autophagy receptors to FIP200 Claw domain for selective autophagy initiation.
Nat Commun, 12, 2021
|
|
3ALM
 
 | | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, mutant C294A | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Kobayashi, J, Yoshida, H, Yoshikane, Y, Kamitori, S, Yagi, T. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
5MR7
 
 | |
7M0E
 
 | | Pre-catalytic synaptic complex of DNA Polymerase Lambda with gapped DSB substrate and incoming dUMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*TP*GP*C)-3'), ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
7B95
 
 | | CooS-V with partially oxidized hybrid cluster by hydroxylamine | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Carbon monoxide dehydrogenase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jeoung, J.H, Dobbek, H. | | Deposit date: | 2020-12-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Morphing [4Fe-3S-nO]-Cluster within a Carbon Monoxide Dehydrogenase Scaffold.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7M09
 
 | | Pre-catalytic quaternary complex of DNA Polymerase Lambda with blunt-ended DSB substrate and incoming dUMPNPP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
