1BSR
 
 | |
4D72
 
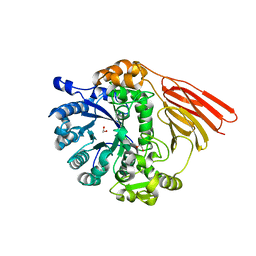 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 2 blood group A-tetrasaccharide (E558A L19 mutant) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, GLYCOSIDE HYDROLASE, ... | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-19 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4HAT
 
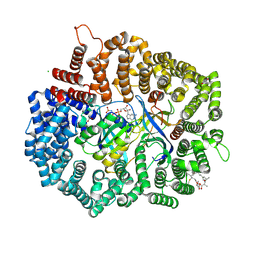 | |
5OTE
 
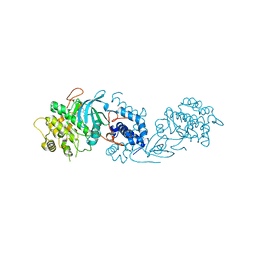 | | MRCK beta in complex with BDP-00008900 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-[(6~{S})-1,8-diazaspiro[5.5]undecan-8-yl]-1~{H}-pyrrolo[2,3-b]pyridin-3-yl]-1,3-thiazole, CHLORIDE ION, ... | | Authors: | Schuettelkopf, A.W. | | Deposit date: | 2017-08-21 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery of Potent and Selective MRCK Inhibitors with Therapeutic Effect on Skin Cancer.
Cancer Res., 78, 2018
|
|
3TGW
 
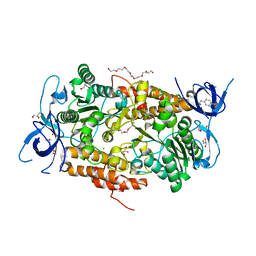 | | Crystal structure of subunit B mutant H156A of the A1AO ATP synthase | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, ... | | Authors: | Tadwal, V.S, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-08-18 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of subunit A mutants H156A, N157A and N157T of the A1AO ATP synthase
To be Published
|
|
1FM4
 
 | | CRYSTAL STRUCTURE OF THE BIRCH POLLEN ALLERGEN BET V 1L | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, MAJOR POLLEN ALLERGEN BET V 1-L | | Authors: | Markovic-Housley, Z, Degano, M, Lamba, D, von Roepenack-Lahaye, E, Clemens, S, Susani, M, Ferreira, F, Scheiner, O, Breiteneder, H. | | Deposit date: | 2000-08-16 | | Release date: | 2002-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of a Hypoallergenic Isoform of the Major Birch Pollen Allergen Bet
v 1 and its Likely Biological Function as a Plant Steroid Carrier
J.Mol.Biol., 325, 2003
|
|
2O6E
 
 | | Structure of native rTp34 from Treponema pallidum from zinc-soaked crystals | | Descriptor: | 1,2-ETHANEDIOL, 34 kDa membrane antigen, CHLORIDE ION, ... | | Authors: | Machius, M, Brautigam, C.A, Deka, R.K, Tomchick, D.R, Lumpkins, S.B, Norgard, M.V. | | Deposit date: | 2006-12-07 | | Release date: | 2006-12-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Tp34 (TP0971) lipoprotein of treponema pallidum: implications of its metal-bound state and affinity for human lactoferrin.
J.Biol.Chem., 282, 2007
|
|
4IWZ
 
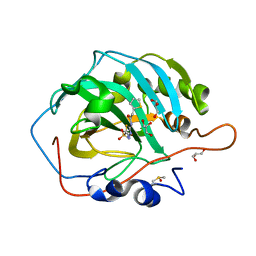 | | structure of hCAII in complex with an acetazolamide derivative | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Biswas, S, McKenna, R. | | Deposit date: | 2013-01-24 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Effect of incorporating a thiophene tail in the scaffold of acetazolamide on the inhibition of human carbonic anhydrase isoforms I, II, IX and XII.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4ESR
 
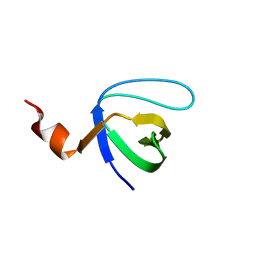 | | Molecular and Structural Characterization of the SH3 Domain of AHI-1 in Regulation of Cellular Resistance of BCR-ABL+ Chronic Myeloid Leukemia Cells to Tyrosine Kinase Inhibitors | | Descriptor: | DI(HYDROXYETHYL)ETHER, Jouberin | | Authors: | Van Petegem, X.F, Liu, P.X, Lobo, P, Jiang, X. | | Deposit date: | 2012-04-23 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Molecular and structural characterization of the SH3 domain of AHI-1 in regulation of cellular resistance of BCR-ABL(+) chronic myeloid leukemia cells to tyrosine kinase inhibitors.
Proteomics, 12, 2012
|
|
2X6E
 
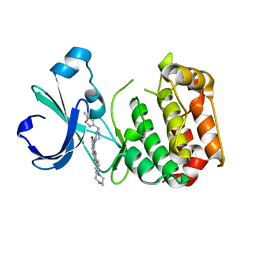 | | Aurora-A bound to an inhibitor | | Descriptor: | 6-BROMO-7-{4-[(5-METHYLISOXAZOL-3-YL)METHYL]PIPERAZIN-1-YL}-2-[4-(4-METHYLPIPERAZIN-1-YL)PHENYL]-1H-IMIDAZO[4,5-B]PYRIDINE, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2010-02-17 | | Release date: | 2010-07-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Imidazo[4,5-b]pyridine derivatives as inhibitors of Aurora kinases: lead optimization studies toward the identification of an orally bioavailable preclinical development candidate.
J. Med. Chem., 53, 2010
|
|
4J0Y
 
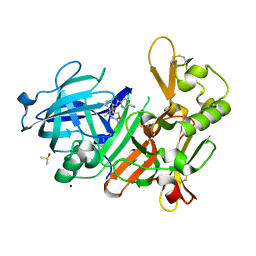 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH 5-Cyano-pyridine-2-carboxylic acid [3-((4R,5S)-2-amino-5-fluoro-4-methyl-5,6-dihydro-4H-[1,3]oxazin-4-yl)-4-fluoro-phenyl]-amide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, N-{3-[(4R,5S)-2-amino-5-fluoro-4-methyl-5,6-dihydro-4H-1,3-oxazin-4-yl]-4-fluorophenyl}-5-cyanopyridine-2-carboxamide, ... | | Authors: | Kuglstatter, A, Stihle, M. | | Deposit date: | 2013-01-31 | | Release date: | 2013-05-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | beta-Secretase (BACE1) Inhibitors with High In Vivo Efficacy Suitable for Clinical Evaluation in Alzheimer s Disease
J.Med.Chem., 56, 2013
|
|
4J1C
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH 5-Cyano-pyridine-2-carboxylic acid [3-((S)-2-amino-5,5-difluoro-4-fluoromethyl-5,6-dihydro-4H-[1,3]oxazin-4-yl)-4-fluoro-phenyl]-amide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, N-{3-[(4S)-2-amino-5,5-difluoro-4-(fluoromethyl)-5,6-dihydro-4H-1,3-oxazin-4-yl]-4-fluorophenyl}-5-cyanopyridine-2-carboxamide, ... | | Authors: | Kuglstatter, A, Stihle, M. | | Deposit date: | 2013-02-01 | | Release date: | 2013-05-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | beta-Secretase (BACE1) Inhibitors with High In Vivo Efficacy Suitable for Clinical Evaluation in Alzheimer s Disease
J.Med.Chem., 56, 2013
|
|
2WQ5
 
 | | Non-antibiotic properties of tetracyclines: structural basis for inhibition of secretory phospholipase A2. | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, CALCIUM ION, PHOSPHOLIPASE A2, ... | | Authors: | Dalm, D, Palm, G.J, Hinrichs, W. | | Deposit date: | 2009-08-13 | | Release date: | 2010-03-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Non-Antibiotic Properties of Tetracyclines: Structural Basis for Inhibition of Secretory Phospholipase A(2).
J.Mol.Biol., 398, 2010
|
|
2XAC
 
 | |
2OCA
 
 | | The crystal structure of T4 UvsW | | Descriptor: | ATP-dependent DNA helicase uvsW | | Authors: | Kerr, I.D, White, S.W. | | Deposit date: | 2006-12-20 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic and NMR Analyses of UvsW and UvsW.1 from Bacteriophage T4.
J.Biol.Chem., 282, 2007
|
|
2WUF
 
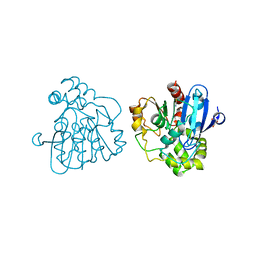 | | Crystal structure of S114A mutant of HsaD from Mycobacterium tuberculosis in complex with 4,9DSHA | | Descriptor: | (3E,5R)-8-[(1S,3AR,4R,7AS)-1-HYDROXY-7A-METHYL-5-OXOOCTAHYDRO-1H-INDEN-4-YL]-5-METHYL-2,6-DIOXOOCT-3-ENOATE, 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE BPHD, GLYCEROL, ... | | Authors: | Lack, N.A, Yam, K.C, Lowe, E.D, Horsman, G.P, Owen, R, Sim, E, Eltis, L.D. | | Deposit date: | 2009-10-02 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of a carbon-carbon hydrolase from Mycobacterium tuberculosis involved in cholesterol metabolism.
J. Biol. Chem., 285, 2010
|
|
1D6Q
 
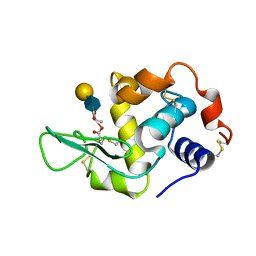 | | HUMAN LYSOZYME E102 MUTANT LABELLED WITH 2',3'-EPOXYPROPYL GLYCOSIDE OF N-ACETYLLACTOSAMINE | | Descriptor: | GLYCEROL, LYSOZYME, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Muraki, M, Harata, K, Sugita, N, Sato, K. | | Deposit date: | 1999-10-15 | | Release date: | 2000-01-21 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Protein-carbohydrate interactions in human lysozyme probed by combining site-directed mutagenesis and affinity labeling.
Biochemistry, 39, 2000
|
|
4AUY
 
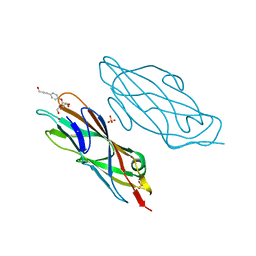 | | Structure of the FimH lectin domain in the trigonal space group, in complex with an hydroxyl propynyl phenyl alpha-D-mannoside at 2.1 A resolution | | Descriptor: | 4-(3-hydroxyprop-1-yn-1-yl)phenyl alpha-D-mannopyranoside, FIMH, NICKEL (II) ION, ... | | Authors: | Wellens, A, Lahmann, M, Touaibia, M, Vaucher, J, Oscarson, S, Roy, R, Remaut, H, Bouckaert, J. | | Deposit date: | 2012-05-22 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | The Tyrosine Gate as a Potential Entropic Lever in the Receptor-Binding Site of the Bacterial Adhesin Fimh.
Biochemistry, 51, 2012
|
|
1O1X
 
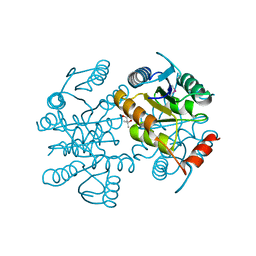 | |
1L6I
 
 | | Crystal Structure of the Maltodextrin Phosphorylase complexed with the products of the enzymatic reaction between glucose-1-phosphate and maltopentaose | | Descriptor: | MALTODEXTRIN PHOSPHORYLASE, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Geremia, S, Campagnolo, M, Schinzel, R, Johnson, L.N. | | Deposit date: | 2002-03-11 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzymatic catalysis in crystals of Escherichia coli maltodextrin phosphorylase
J.Mol.Biol., 322, 2002
|
|
4J17
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH 5-Cyano-pyridine-2-carboxylic acid [3-((S)-2-amino-4-difluoromethyl-5,6-dihydro-4H-[1,3]oxazin-4-yl)-4-fluoro-phenyl]-amide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, N-{3-[(4S)-2-amino-4-(difluoromethyl)-5,6-dihydro-4H-1,3-oxazin-4-yl]-4-fluorophenyl}-5-cyanopyridine-2-carboxamide, ... | | Authors: | Kuglstatter, A, Stihle, M. | | Deposit date: | 2013-02-01 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | beta-Secretase (BACE1) Inhibitors with High In Vivo Efficacy Suitable for Clinical Evaluation in Alzheimer s Disease
J.Med.Chem., 56, 2013
|
|
5REX
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102287 | | Descriptor: | 1-{4-[(naphthalen-1-yl)methyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
1FQY
 
 | | STRUCTURE OF AQUAPORIN-1 AT 3.8 A RESOLUTION BY ELECTRON CRYSTALLOGRAPHY | | Descriptor: | AQUAPORIN-1 | | Authors: | Murata, K, Mitsuoka, K, Hirai, T, Walz, T, Agre, P, Heymann, J.B, Engel, A, Fujiyoshi, Y. | | Deposit date: | 2000-09-07 | | Release date: | 2000-10-18 | | Last modified: | 2024-04-17 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.8 Å) | | Cite: | Structural determinants of water permeation through aquaporin-1.
Nature, 407, 2000
|
|
1XON
 
 | | Catalytic Domain Of Human Phosphodiesterase 4D In Complex With Piclamilast | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(CYCLOPENTYLOXY)-N-(3,5-DICHLOROPYRIDIN-4-YL)-4-METHOXYBENZAMIDE, ... | | Authors: | Card, G.L, England, B.P, Suzuki, Y, Fong, D, Powell, B, Lee, B, Luu, C, Tabrizizad, M, Gillette, S, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-10-06 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Basis for the Activity of Drugs that Inhibit Phosphodiesterases.
STRUCTURE, 12, 2004
|
|
3TY1
 
 | |
