2XCK
 
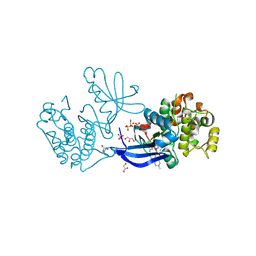 | | Crystal structure of PDK1 in complex with a pyrazoloquinazoline inhibitor | | Descriptor: | 1-METHYL-8-{[4-(4-METHYLPIPERAZIN-1-YL)PHENYL]AMINO}-N-[(2-METHYLPYRIDIN-4-YL)METHYL]-4,5-DIHYDRO-1H-PYRAZOLO[4,3-H]QUINAZOLINE-3-CARBOXAMIDE, 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE 1, GLYCEROL, ... | | Authors: | Angiolini, M, Banfi, P, Casale, E, Casuscelli, F, Fiorelli, C, Saccardo, M.B, Silvagni, M, Zuccotto, F. | | Deposit date: | 2010-04-23 | | Release date: | 2010-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Optimization of Potent Pdk1 Inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7GTC
 
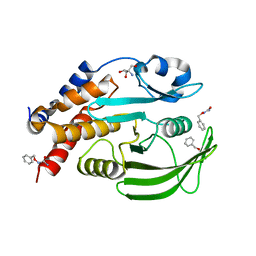 | |
3VJS
 
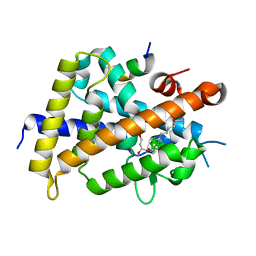 | | Vitamin D receptor complex with a carborane compound | | Descriptor: | 1-(2-[(S)-2,4-Dihydroxybutoxy]ethyl)-12-(5-ethyl-5-hydroxyheptyl)-1,12-dicarba-closo-dodecaborane, Vitamin D3 receptor, peptide from Mediator of RNA polymerase II transcription subunit 1 | | Authors: | Fujii, S, Masuno, M, Kagechika, H, Nakabayashi, M, Ito, N. | | Deposit date: | 2011-10-31 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Boron Cluster-based Development of Potent Nonsecosteroidal Vitamin D Receptor Ligands: Direct Observation of Hydrophobic Interaction between Protein Surface and Carborane
J.Am.Chem.Soc., 133, 2011
|
|
3MWO
 
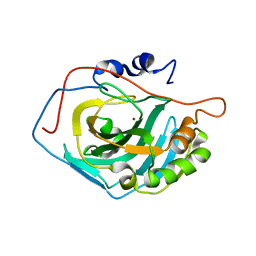 | |
2C2O
 
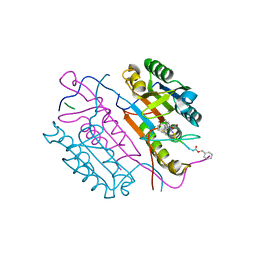 | | Crystal structures of caspase-3 in complex with aza-peptide Michael acceptor inhibitors. | | Descriptor: | AZA-PEPTIDE INHIBITOR (5S, 8R, 11S)-14-{4-[BENZYL(METHYL) AMINO]-4-OXOBUTANOYL}-8-(2-CARBOXYETHYL)-5-(CARBOXYMETHYL)-11-(1-METHYLETHYL)-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,13,14-PENTAAZAHEXADECAN-16-OIC ACID, ... | | Authors: | Ganesan, R, Jelakovic, S, Ekici, O.D, Li, Z.Z, James, K.E, Asgian, J.L, Campbell, A, Mikolajczyk, J, Salvesen, G.S, Gruetter, M.G, Powers, J.C. | | Deposit date: | 2005-09-29 | | Release date: | 2006-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Design, Synthesis, and Evaluation of Aza-Peptide Michael Acceptors as Selective and Potent Inhibitors of Caspases-2, -3, -6, -7, -8, -9, and - 10.
J.Med.Chem., 49, 2006
|
|
3VTP
 
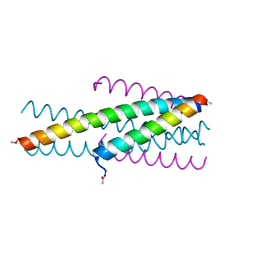 | | HIV fusion inhibitor MT-C34 | | Descriptor: | Transmembrane protein gp41 | | Authors: | Yao, X, Waltersperger, S, Wang, M, Cui, S. | | Deposit date: | 2012-06-02 | | Release date: | 2012-08-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The M-T hook structure is critical for design of HIV-1 fusion inhibitors.
J.Biol.Chem., 287, 2012
|
|
1U68
 
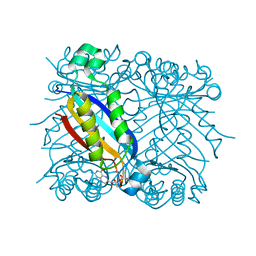 | | DHNA 7,8 DIHYDRONEOPTERIN COMPLEX | | Descriptor: | 2-AMINO-7,8-DIHYDRO-6-(1,2,3-TRIHYDROXYPROPYL)-4(1H)-PTERIDINONE, Dihydroneopterin aldolase | | Authors: | Sanders, W.J, Nienaber, V.L, Lerner, C.G, McCall, J.O, Merrick, S.M, Swanson, S.J, Harlan, J.E, Stoll, V.S, Stamper, G.F, Betz, S.F, Condroski, K.R, Meadows, R.P, Severin, J.M, Walter, K.A, Magdalinos, P, Jakob, C.G, Wagner, R, Beutel, B.A. | | Deposit date: | 2004-07-29 | | Release date: | 2004-10-19 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of potent inhibitors of dihydroneopterin aldolase using CrystaLEAD high-throughput X-ray crystallographic screening and structure-directed lead optimization.
J.MED.CHEM., 47, 2004
|
|
4IEJ
 
 | |
3G5R
 
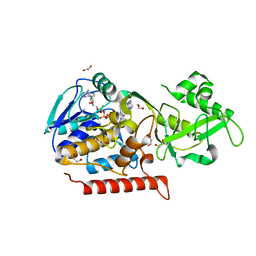 | | Crystal structure of Thermus thermophilus TrmFO in complex with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Nishimasu, H, Ishitani, R, Hori, H, Nureki, O. | | Deposit date: | 2009-02-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atomic structure of a folate/FAD-dependent tRNA T54 methyltransferase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3VUG
 
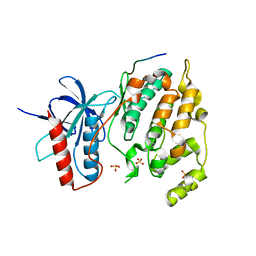 | | Crystal structure of a cysteine-deficient mutant M2 in MAP kinase JNK1 | | Descriptor: | Mitogen-activated protein kinase 8, Peptide from C-Jun-amino-terminal kinase-interacting protein 1, SULFATE ION | | Authors: | Nakaniwa, T, Kinoshita, T, Inoue, T. | | Deposit date: | 2012-06-28 | | Release date: | 2013-02-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Seven cysteine-deficient mutants depict the interplay between thermal and chemical stabilities of individual cysteine residues in mitogen-activated protein kinase c-Jun N-terminal kinase 1
Biochemistry, 51, 2012
|
|
3T6C
 
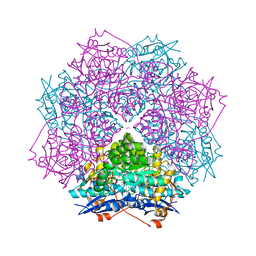 | | Crystal structure of an enolase from pantoea ananatis (efi target efi-501676) with bound d-gluconate and mg | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, D-gluconic acid, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-07-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Crystal structure of an enolase from pantoea ananatis (efi target efi-501676) with bound d-gluconate and mg
to be published
|
|
2EBU
 
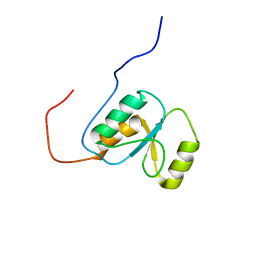 | |
2NVN
 
 | |
2G96
 
 | |
3VII
 
 | | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with Bis-Tris | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase, GLYCEROL, ... | | Authors: | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | High-resolution structures of Neotermes koshunensis beta-glucosidase mutants provide insights into the catalytic mechanism and the synthesis of glucoconjugates
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3NRE
 
 | |
4PJD
 
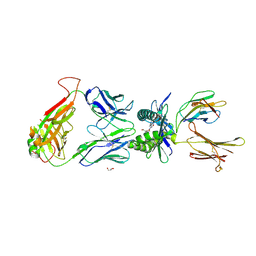 | | Structure of human MR1-5-OP-RU in complex with human MAIT C-C10 TCR | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2014-05-12 | | Release date: | 2014-07-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells.
J.Exp.Med., 211, 2014
|
|
2B8W
 
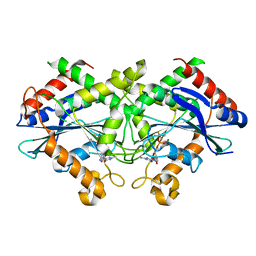 | | Crystal-structure of the N-terminal Large GTPase Domain of human Guanylate Binding protein 1 (hGBP1) in complex with GMP/AlF4 | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Interferon-induced guanylate-binding protein 1, MAGNESIUM ION, ... | | Authors: | Ghosh, A, Praefcke, G.J.K, Renault, L, Wittinghofer, A, Herrmann, C. | | Deposit date: | 2005-10-10 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP.
Nature, 440, 2006
|
|
2R9B
 
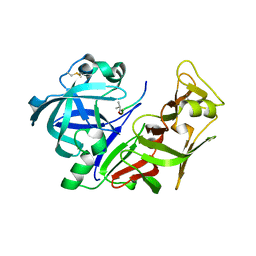 | | Structural Analysis of Plasmepsin 2 from Plasmodium falciparum complexed with a peptide-based inhibitor | | Descriptor: | Plasmepsin-2, peptide-based inhibitor | | Authors: | Liu, P, Marzahn, M.R, Robbins, A.H, McKenna, R, Dunn, B.M. | | Deposit date: | 2007-09-12 | | Release date: | 2007-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recombinant plasmepsin 1 from the human malaria parasite plasmodium falciparum: enzymatic characterization, active site inhibitor design, and structural analysis.
Biochemistry, 48, 2009
|
|
2B77
 
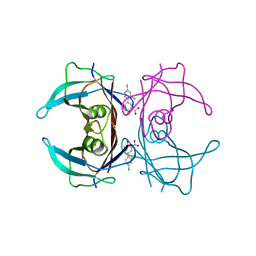 | | Human transthyretin (TTR) complexed with Diflunisal analogues- TTR.2',4'-DICHLORO-4-HYDROXY-1,1'-BIPHENYL-3-CARBOXYLIC ACID | | Descriptor: | 2',4'-DICHLORO-4-HYDROXY-1,1'-BIPHENYL-3-CARBOXYLIC ACID, Transthyretin | | Authors: | Palaninathan, S.K, Kelly, J.W, Sacchettini, J.C. | | Deposit date: | 2005-10-03 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Diflunisal analogues stabilize the native state of transthyretin. Potent inhibition of amyloidogenesis.
J.Med.Chem., 47, 2004
|
|
3GB0
 
 | |
3GBR
 
 | | Anthranilate phosphoribosyl-transferase (TRPD) double mutant D83G F149S from S. solfataricus | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Anthranilate phosphoribosyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bruning, M, Schlee, S, Deuss, M, Ivens, A, Mayans, O, Sterner, R. | | Deposit date: | 2009-02-20 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Activation of anthranilate phosphoribosyltransferase from Sulfolobus solfataricus by removal of magnesium inhibition and acceleration of product release
Biochemistry, 48, 2009
|
|
2XUU
 
 | | Crystal structure of a DAP-kinase 1 mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DEATH-ASSOCIATED PROTEIN KINASE 1, MAGNESIUM ION, ... | | Authors: | de Diego, I, Kuper, J, Lehmann, F, Wilmanns, M. | | Deposit date: | 2010-10-21 | | Release date: | 2011-11-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Pef/Y Substrate Recognition and Signature Motif Plays a Critical Role in Dapk-Related Kinase Activity.
Chem.Biol., 21, 2014
|
|
4F2G
 
 | | The Crystal Structure of Ornithine carbamoyltransferase from Burkholderia thailandensis E264 | | Descriptor: | 1,2-ETHANEDIOL, Ornithine carbamoyltransferase 1, PHOSPHATE ION | | Authors: | Craig, T.K, Fox, D, Staker, B, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2012-05-07 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Combining functional and structural genomics to sample the essential Burkholderia structome.
Plos One, 8, 2013
|
|
3R9P
 
 | |
