5IED
 
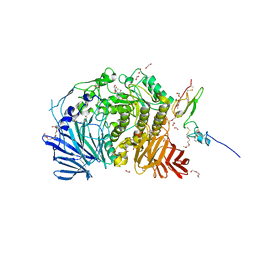 | | Murine endoplasmic reticulum alpha-glucosidase II with castanospermine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2016-02-25 | | Release date: | 2016-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
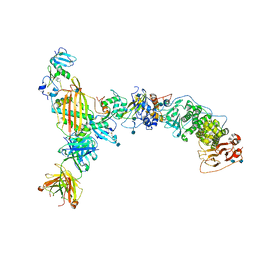
5VOB
 
 | |
5VPD
 
 | | Transcription factor FosB/JunD bZIP domain in its oxidized form, type-III crystal | | Descriptor: | CHLORIDE ION, Protein fosB, SODIUM ION, ... | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
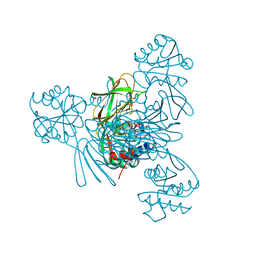
7MH7
 
 | |
6CX5
 
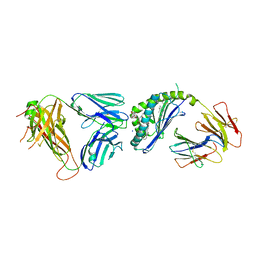 | | Structure of alpha-GSA[8,8P] bound by CD1d and in complex with the Va14Vb8.2 TCR | | Descriptor: | (5R,6S,7S)-5,6-dihydroxy-7-(octanoylamino)-N-(8-phenyloctyl)-8-{[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]oxy}octanamide (non-preferred name), 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, J, Zajonc, D. | | Deposit date: | 2018-04-02 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
5IH7
 
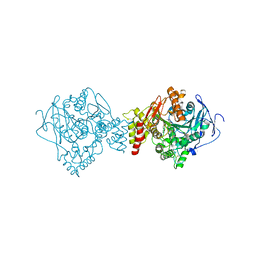 | |
7MJJ
 
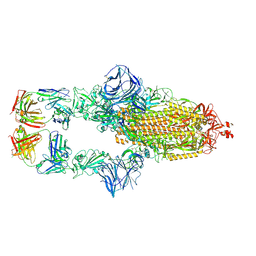 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to Fab ab1 (class 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ab1 Heavy Chain, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
5IIC
 
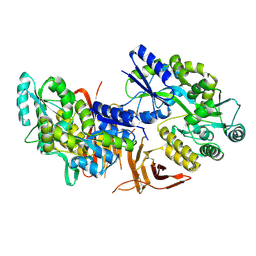 | | Crystal structure of red abalone VERL repeat 3 at 2.9 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Maltose-binding periplasmic protein,Vitelline envelope sperm lysin receptor, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Sadat Al-Hosseini, H, Raj, I, Nishimura, K, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
7MKL
 
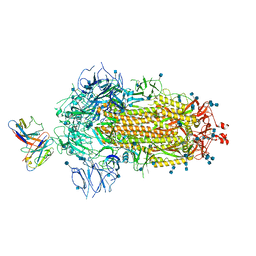 | |
5IK8
 
 | | Laminin A2LG45 I-form, G6/7 bound. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-hydroxy-4-methyl-2H-chromen-2-one, ... | | Authors: | Briggs, D.C, Hohenester, E, Campbell, K.P. | | Deposit date: | 2016-03-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of laminin binding to the LARGE glycans on dystroglycan.
Nat.Chem.Biol., 12, 2016
|
|
5VLI
 
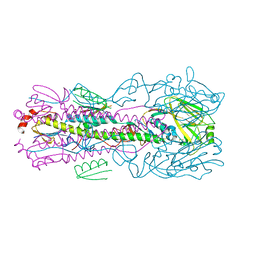 | | Computationally designed inhibitor peptide HB1.6928.2.3 in complex with influenza hemagglutinin (A/PuertoRico/8/1934) | | Descriptor: | 2,5,8,11-TETRAOXATRIDECANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bernard, S.M, Wilson, I.A. | | Deposit date: | 2017-04-25 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Massively parallel de novo protein design for targeted therapeutics.
Nature, 550, 2017
|
|
7MJH
 
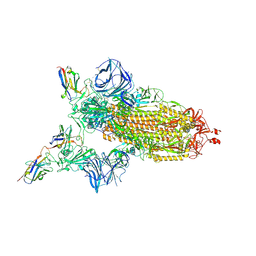 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to VH ab8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
5IKT
 
 | | The Structure of Tolfenamic Acid Bound to Human Cyclooxygenase-2 | | Descriptor: | 2-[(3-chloro-2-methylphenyl)amino]benzoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Orlando, B.J, Malkowski, M.G. | | Deposit date: | 2016-03-03 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Substrate-selective Inhibition of Cyclooxygeanse-2 by Fenamic Acid Derivatives Is Dependent on Peroxide Tone.
J.Biol.Chem., 291, 2016
|
|
6CW9
 
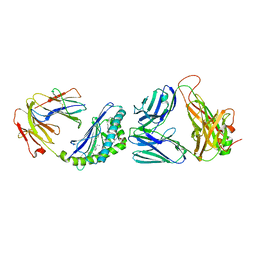 | | Structure of alpha-GC[8,16P] bound by CD1d and in complex with the Va14Vb8.2 TCR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, J, Zajonc, D. | | Deposit date: | 2018-03-30 | | Release date: | 2019-04-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
5VPG
 
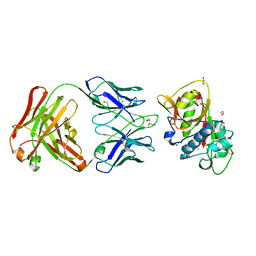 | | CRYSTAL STRUCTURE OF DER P 1 COMPLEXED WITH FAB 4C1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Chruszcz, M, Vailes, L.D, Chapman, M.D, Pomes, A, Minor, W. | | Deposit date: | 2017-05-05 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Determinants For Antibody Binding On Group 1 House Dust Mite Allergens.
J.Biol.Chem., 287, 2012
|
|
7MMO
 
 | | LY-CoV1404 neutralizing antibody against SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LY-CoV1404 Fab heavy chain, LY-CoV1404 Fab light chain, ... | | Authors: | Hendle, J, Pustilnik, A, Sauder, J.M, Coleman, K.A, Boyles, J.S, Dickinson, C.D. | | Deposit date: | 2021-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.427 Å) | | Cite: | LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants.
Biorxiv, 2022
|
|
7MJK
 
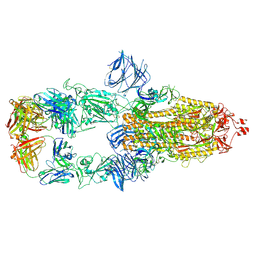 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to Fab ab1 (class 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ab1 Heavy Chain, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJG
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJM
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to human ACE2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
6D24
 
 | | Trypanosoma cruzi Glucose-6-P Dehydrogenase in complex with G6P | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Botti, H, Ortiz, C, Comini, M.A, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2018-04-12 | | Release date: | 2018-05-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Glucose-6-Phosphate Dehydrogenase from the Human Pathogen Trypanosoma cruzi Evolved Unique Structural Features to Support Efficient Product Formation.
J.Mol.Biol., 431, 2019
|
|
7MOA
 
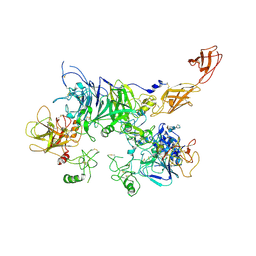 | | Cryo-EM structure of the c-MET II/HGF I complex bound with HGF II in a rigid conformation | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
6CX7
 
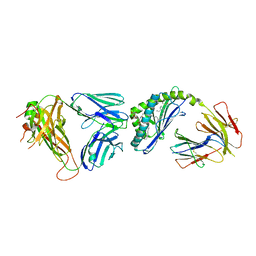 | | Structure of alpha-GSA[12,6P] bound by CD1d and in complex with the Va14Vb8.2 TCR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Wang, J, Zajonc, D. | | Deposit date: | 2018-04-02 | | Release date: | 2019-04-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
7N0H
 
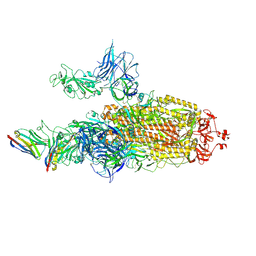 | | CryoEM structure of SARS-CoV-2 spike protein (S-6P, 2-up) in complex with sybodies (Sb45) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Jiang, J, Huang, R, Margulies, D. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
5VMG
 
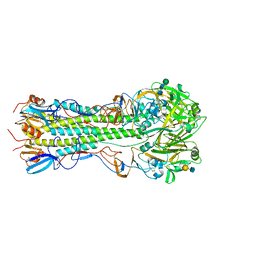 | | Influenza hemagglutinin H1 mutant DH1E in complex with 6'SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Ni, F, Kondrashkina, E, Wang, Q. | | Deposit date: | 2017-04-27 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Determinant of receptor-preference switch in influenza hemagglutinin.
Virology, 513, 2017
|
|
7N0G
 
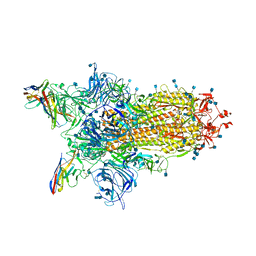 | | CryoEm structure of SARS-CoV-2 spike protein (S-6P, 1-up) in complex with sybodies (Sb45) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Jiang, J, Huang, R, Margulies, D. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
