7A8M
 
 | |
6KII
 
 | | photolyase from Arthrospira platensis | | Descriptor: | 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, Deoxyribopyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yan, H, Zhu, K. | | Deposit date: | 2019-07-18 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A newly identified photolyase from Arthrospira platensis possesses a unique methenyltetrahydrofolate chromophore-binding pattern.
Febs Lett., 594, 2020
|
|
8AAB
 
 | | S148F mutant of blue-to-red fluorescent timer mRubyFT | | Descriptor: | mRubyFT S148F mutant of blue-to-red fluorescent timer | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Dorovatovskii, P.V, Subach, O.M, Popov, V.O, Subach, F.V. | | Deposit date: | 2022-06-30 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | mRubyFT/S147I, a mutant of blue-to-red fluorescent timer
Crystallography Reports, 2022
|
|
2HGD
 
 | |
8YLZ
 
 | | Structure of a cis-Geranylfarnesyl Diphosphate Synthase from Streptomyces clavuligerus | | Descriptor: | Isoprenyl transferase | | Authors: | Li, F.R, Wang, Q.L, Pan, X.M, Dong, L.B. | | Deposit date: | 2024-03-07 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery, Structure, and Engineering of a cis-Geranylfarnesyl Diphosphate Synthase.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
6S36
 
 | | Crystal structure of E. coli Adenylate kinase R119K mutant | | Descriptor: | Adenylate kinase, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Grundstrom, C, Rogne, P, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2019-06-24 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nucleation of an Activating Conformational Change by a Cation-pi Interaction.
Biochemistry, 58, 2019
|
|
6YLM
 
 | | mCherry | | Descriptor: | CHLORIDE ION, mCherry | | Authors: | Myskova, J, Rybakova, M, Brynda, J, Lazar, J. | | Deposit date: | 2020-04-07 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Directionality of light absorption and emission in representative fluorescent proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4DXI
 
 | |
5Y00
 
 | | Acid-tolerant monomeric GFP, Gamillus, fluorescence (ON) state | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Nakashima, R, Sakurai, K, Shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2017-07-14 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Acid-Tolerant Monomeric GFP from Olindias formosa.
Cell Chem Biol, 25, 2018
|
|
5T3I
 
 | |
1Q73
 
 | |
3DQ2
 
 | |
3DQ6
 
 | |
4KEX
 
 | |
6QSL
 
 | |
6RUM
 
 | | Crystal structure of GFP-LAMA-G97 - a GFP enhancer nanobody with cpDHFR insertion and TMP and NADPH | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GFP-LAMA-G97 a GFP enhancer nanobody with cpDHFR insertion, ... | | Authors: | Farrants, H, Tarnawski, M, Mueller, T.G, Otsuka, S, Hiblot, J, Koch, B, Kueblbeck, M, Kraeusslich, H.-G, Ellenberg, J, Johnsson, K. | | Deposit date: | 2019-05-28 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chemogenetic Control of Nanobodies.
Nat.Methods, 17, 2020
|
|
4LQU
 
 | | 1.60A resolution crystal structure of a superfolder green fluorescent protein (W57G) mutant | | Descriptor: | Green fluorescent protein | | Authors: | Lovell, S, Xia, Y, Vo, B, Battaile, K.P, Egan, C, Karanicolas, J. | | Deposit date: | 2013-07-19 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The designability of protein switches by chemical rescue of structure: mechanisms of inactivation and reactivation.
J.Am.Chem.Soc., 135, 2013
|
|
7ODC
 
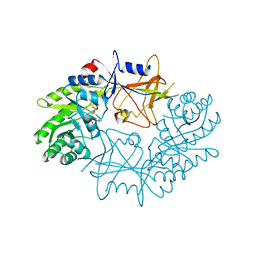 | | CRYSTAL STRUCTURE ORNITHINE DECARBOXYLASE FROM MOUSE, TRUNCATED 37 RESIDUES FROM THE C-TERMINUS, TO 1.6 ANGSTROM RESOLUTION | | Descriptor: | PROTEIN (ORNITHINE DECARBOXYLASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Kern, A.D, Oliveira, M.A, Coffino, P, Hackert, M.L. | | Deposit date: | 1999-03-03 | | Release date: | 1999-10-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of mammalian ornithine decarboxylase at 1.6 A resolution: stereochemical implications of PLP-dependent amino acid decarboxylases.
Structure Fold.Des., 7, 1999
|
|
3AI4
 
 | | Crystal structure of yeast enhanced green fluorescent protein - mouse polymerase iota ubiquitin binding motif fusion protein | | Descriptor: | SULFATE ION, yeast enhanced green fluorescent protein,DNA polymerase iota | | Authors: | Suzuki, N, Wakatsuki, S, Kawasaki, M. | | Deposit date: | 2010-05-10 | | Release date: | 2010-09-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallization of small proteins assisted by green fluorescent protein
Acta Crystallogr.,Sect.D, 66, 2010
|
|
7SAH
 
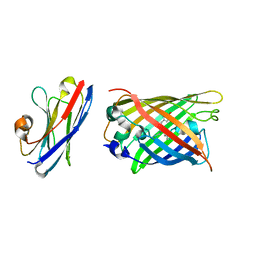 | |
1ZIN
 
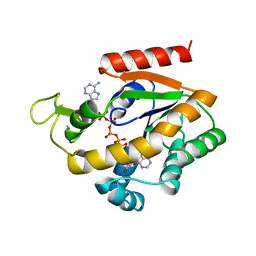 | | ADENYLATE KINASE WITH BOUND AP5A | | Descriptor: | ADENYLATE KINASE, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ZINC ION | | Authors: | Berry, M.B, Phillips Jr, G.N. | | Deposit date: | 1996-06-07 | | Release date: | 1997-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of Bacillus stearothermophilus adenylate kinase with bound Ap5A, Mg2+ Ap5A, and Mn2+ Ap5A reveal an intermediate lid position and six coordinate octahedral geometry for bound Mg2+ and Mn2+.
Proteins, 32, 1998
|
|
3WUP
 
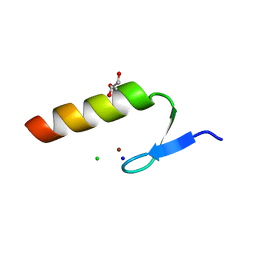 | | Crystal Structure of the Ubiquitin-Binding Zinc Finger (UBZ) Domain of the Human DNA Polymerase Eta | | Descriptor: | CHLORIDE ION, DNA polymerase eta, GLYCEROL, ... | | Authors: | Suzuki, N, Wakatsuki, S, Kawasaki, S. | | Deposit date: | 2014-05-01 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A novel mode of ubiquitin recognition by the ubiquitin-binding zinc finger domain of WRNIP1.
Febs J., 283, 2016
|
|
7Z7Q
 
 | |
3GEX
 
 | |
5AZB
 
 | | Crystal structure of Escherichia coli Lgt in complex with phosphatidylglycerol and the inhibitor palmitic acid | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, PALMITIC ACID, Prolipoprotein diacylglyceryl transferase, ... | | Authors: | Zhang, X.C, Mao, G, Zhao, Y. | | Deposit date: | 2015-09-30 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of E. coli lipoprotein diacylglyceryl transferase
Nat Commun, 7, 2016
|
|
