4YE1
 
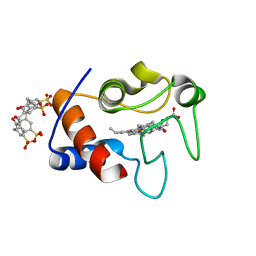 | | A cytochrome c plus calixarene structure - alternative ligand binding mode | | Descriptor: | 25,26,27,28-tetrahydroxypentacyclo[19.3.1.1~3,7~.1~9,13~.1~15,19~]octacosa-1(25),3(28),4,6,9(27),10,12,15(26),16,18,21,23-dodecaene-5,11,17,23-tetrasulfonic acid, Cytochrome c iso-1, GLYCEROL, ... | | Authors: | Mallon, M.M, McGovern, R.E, McCarty, A.A, Crowley, P.B. | | Deposit date: | 2015-02-23 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A cytochrome c-calixarene structure
To Be Published
|
|
6GJI
 
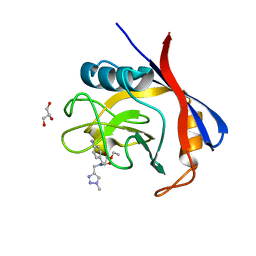 | | Cyclophilin A complexed with the tri-vector ligand 8. | | Descriptor: | GLYCEROL, Peptidyl-prolyl cis-trans isomerase A, ethyl 2-[[(4-aminophenyl)methyl-[(1-methyl-1,2,3-triazol-4-yl)methyl]carbamoyl]amino]ethanoate | | Authors: | Georgiou, C, De Simone, A, Walkinshaw, M.D, Michel, J. | | Deposit date: | 2018-05-16 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A computationally designed binding mode flip leads to a novel class of potent tri-vector cyclophilin inhibitors.
Chem Sci, 10, 2019
|
|
6GNH
 
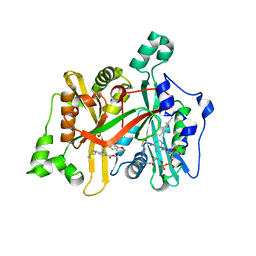 | | Crystal Structure of Leishmania major N-Myristoyltransferase (NMT) With Bound Myristoyl-CoA and an Azepanyl Phenyl Benzylsulphonamide Ligand | | Descriptor: | Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA, methyl 4-(azepan-1-yl)-3-[(4-methoxyphenyl)sulfonylamino]benzoate | | Authors: | Robinson, D.A, Harrison, J.R, Brand, S, Smith, V.C, Thompson, S, Smith, A, Davies, K, Mok, N.Y, Torrie, L.S, Collie, I, Hallyburton, I, Norval, S, Simeons, F.R.C, Stojanovski, L, Frearson, J.A, Brenk, R, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2018-05-30 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Molecular Hybridization Approach for the Design of Potent, Highly Selective, and Brain-Penetrant N-Myristoyltransferase Inhibitors.
J. Med. Chem., 61, 2018
|
|
6GJM
 
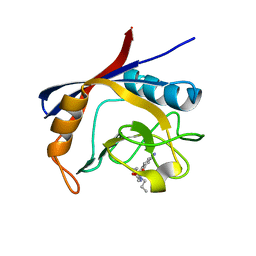 | | Cyclophilin A complexed with tri-vector ligand 4. | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, ethyl 2-[[(4-aminophenyl)methyl-propyl-carbamoyl]amino]ethanoate | | Authors: | Georgiou, C, De Simone, A, Walkinshaw, M.D, Michel, J. | | Deposit date: | 2018-05-16 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.354 Å) | | Cite: | A computationally designed binding mode flip leads to a novel class of potent tri-vector cyclophilin inhibitors.
Chem Sci, 10, 2019
|
|
6CUH
 
 | | Crystal structure of the unliganded BC8B TCR | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shahine, A.E, Rossjohn, J. | | Deposit date: | 2018-03-26 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A T-cell receptor escape channel allows broad T-cell response to CD1b and membrane phospholipids.
Nat Commun, 10, 2019
|
|
7F5D
 
 | | Crystal structure of BPTF-BRD with ligand DC-BPi-03 bound | | Descriptor: | 6-(1H-indol-5-yl)-N-methyl-2-methylsulfonyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.57150865 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
5AB2
 
 | | Crystal structure of aminopeptidase ERAP2 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mpakali, A, Giastas, P, Saridakis, E, Mavridis, I.M, Stratikos, E. | | Deposit date: | 2015-07-31 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.729 Å) | | Cite: | Structural Basis for Antigenic Peptide Recognition and Processing by Endoplasmic Reticulum (Er) Aminopeptidase 2.
J.Biol.Chem., 290, 2015
|
|
7F5C
 
 | | Crystal structure of BPTF-BRD with ligand DC-BPi-07 bound | | Descriptor: | 6-[1-[3-(dimethylamino)propyl]indol-5-yl]-2-methylsulfonyl-N-propyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65004492 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
7F5E
 
 | | Crystal structure of BPTF-BRD with ligand DC-BPi-11 bound | | Descriptor: | N,N-dimethyl-3-[5-(2-methylsulfonyl-7H-pyrrolo[2,3-d]pyrimidin-4-yl)indol-1-yl]propan-1-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.20017123 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
2GX1
 
 | | Solution structure and alanine scan of a spider toxin that affects the activation of mammalian sodium channels | | Descriptor: | Neurotoxin magi-5 | | Authors: | Sabo, J.K, Corzo, G, Bosmans, F, Billen, B, Villegas, E, Tytgat, J, Norton, R.S. | | Deposit date: | 2006-05-08 | | Release date: | 2006-12-05 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Alanine Scan of a Spider Toxin That Affects the Activation of Mammalian Voltage-gated Sodium Channels
J.Biol.Chem., 282, 2007
|
|
3DZ6
 
 | | Human AdoMetDC with 5'-[(4-aminooxybutyl)methylamino]-5'deoxy-8-ethyladenosine | | Descriptor: | 1,4-DIAMINOBUTANE, 5'-{[4-(aminooxy)butyl](methyl)amino}-5'-deoxy-8-ethenyladenosine, S-adenosylmethionine decarboxylase alpha chain, ... | | Authors: | Bale, S, McCloskey, D.E, Pegg, A.E, Secrist III, J.A, Guida, W.C, Ealick, S.E. | | Deposit date: | 2008-07-29 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | New Insights into the Design of Inhibitors of Human S-Adenosylmethionine Decarboxylase: Studies of Adenine C8 Substitution in Structural Analogues of S-Adenosylmethionine
J.Med.Chem., 52, 2009
|
|
3MBA
 
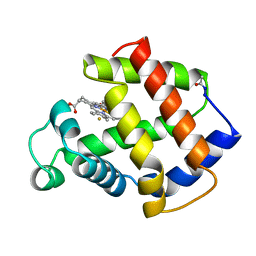 | | APLYSIA LIMACINA MYOGLOBIN. CRYSTALLOGRAPHIC ANALYSIS AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | FLUORIDE ION, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bolognesi, M, Onesti, S, Gatti, G, Coda, A, Ascenzi, P, Brunori, M. | | Deposit date: | 1989-02-22 | | Release date: | 1990-01-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aplysia limacina myoglobin. Crystallographic analysis at 1.6 A resolution.
J.Mol.Biol., 205, 1989
|
|
1O6G
 
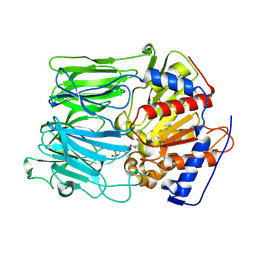 | |
3MTX
 
 | | Crystal structure of chicken MD-1 | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, GLYCEROL, Protein MD-1, ... | | Authors: | Yoon, S.I, Hong, M, Han, G.W, Wilson, I.A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-06-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of soluble MD-1 and its interaction with lipid IVa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6JT0
 
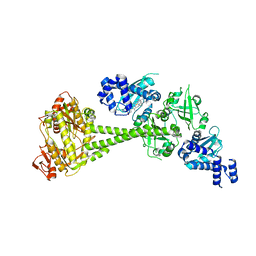 | | Structure of human soluble guanylate cyclase in the unliganded state | | Descriptor: | Guanylate cyclase soluble subunit alpha-1, Guanylate cyclase soluble subunit beta-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chen, L, Kang, Y, Liu, R, Wu, J.-X. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the mechanism of human soluble guanylate cyclase.
Nature, 574, 2019
|
|
4EJG
 
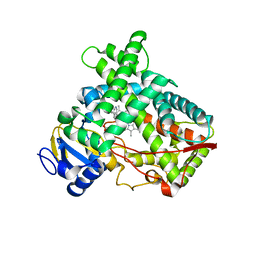 | | Human Cytochrome P450 2A13 in complex with Nicotine | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, Cytochrome P450 2A13, GLYCEROL, ... | | Authors: | DeVore, N.M, Scott, E.E. | | Deposit date: | 2012-04-06 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nicotine and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone binding and access channel in human cytochrome P450 2A6 and 2A13 enzymes.
J.Biol.Chem., 287, 2012
|
|
2GVX
 
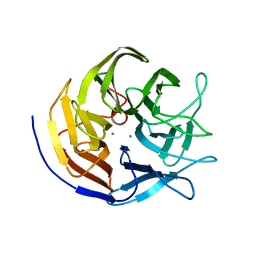 | | Structure of diisopropyl fluorophosphatase (DFPase), mutant D229N / N175D | | Descriptor: | CALCIUM ION, diisopropyl fluorophosphatase | | Authors: | Blum, M.-M, Lohr, F, Richardt, A, Ruterjans, H, Chen, J.C.-H. | | Deposit date: | 2006-05-03 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of a Designed Substrate Analogue to Diisopropyl Fluorophosphatase: Implications for the Phosphotriesterase Mechanism
J.Am.Chem.Soc., 128, 2006
|
|
6JYU
 
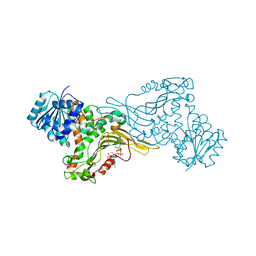 | | Crystal structure of Human G6PD Canton | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Au, S.W.N. | | Deposit date: | 2019-04-28 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of Human G6PD Canton
To Be Published
|
|
2MCG
 
 | |
7C88
 
 | | Complex structure of JS003 and PD-L1 | | Descriptor: | JS003 Heavy chain, JS003 Light chain, Programmed cell death 1 ligand 1 | | Authors: | Bi, X, Shi, R, Chai, Y, Qi, J, Yan, J, Tan, S. | | Deposit date: | 2020-05-29 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Identification of a hotspot on PD-L1 for pH-dependent binding by monoclonal antibodies for tumor therapy.
Signal Transduct Target Ther, 5, 2020
|
|
2HBC
 
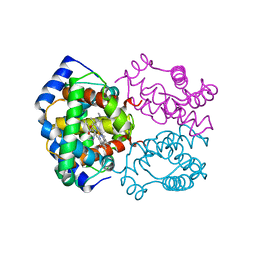 | | HIGH RESOLUTION X-RAY STRUCTURES OF MYOGLOBIN-AND HEMOGLOBIN-ALKYL ISOCYANIDE COMPLEXES | | Descriptor: | ETHYL ISOCYANIDE, HEMOGLOBIN A (ETHYL ISOCYANIDE) (ALPHA CHAIN), HEMOGLOBIN A (ETHYL ISOCYANIDE) (BETA CHAIN), ... | | Authors: | Johnson, K.A, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1994-08-31 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High Resolution X-Ray Structures of Myoglobin-and Hemoglobin-Alkyl Isocyanide Complexes
Thesis, 1, 1993
|
|
7FEW
 
 | |
7FF8
 
 | |
2KI5
 
 | | HERPES SIMPLEX TYPE-1 THYMIDINE KINASE IN COMPLEX WITH THE DRUG ACICLOVIR AT 1.9A RESOLUTION | | Descriptor: | 9-HYROXYETHOXYMETHYLGUANINE, PROTEIN (THYMIDINE KINASE), SULFATE ION | | Authors: | Bennett, M.S, Wien, F, Champness, J.N, Batuwangala, T, Rutherford, T, Summers, W.C, Sun, H, Wright, G, Sanderson, M.R. | | Deposit date: | 1999-02-12 | | Release date: | 1999-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure to 1.9 A resolution of a complex with herpes simplex virus type-1 thymidine kinase of a novel, non-substrate inhibitor: X-ray crystallographic comparison with binding of aciclovir.
FEBS Lett., 443, 1999
|
|
7FF9
 
 | |
