1MD8
 
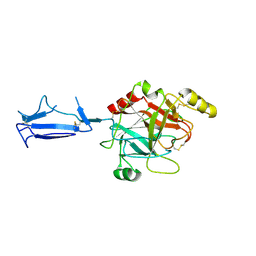 | | Monomeric structure of the active catalytic domain of complement protease C1r | | Descriptor: | C1R COMPLEMENT SERINE PROTEASE | | Authors: | Budayova-Spano, M, Grabarse, W, Thielens, N.M, Hillen, H, Lacroix, M, Schmidt, M, Fontecilla-Camps, J, Arlaud, G.J, Gaboriaud, C. | | Deposit date: | 2002-08-07 | | Release date: | 2003-08-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Monomeric structures of the zymogen and active catalytic domain of complement protease c1r: further insights into the c1 activation mechanism
Structure, 10, 2002
|
|
1L4Z
 
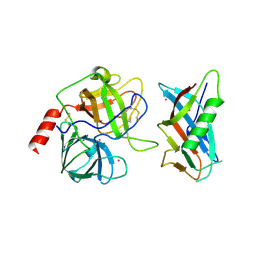 | | X-RAY CRYSTAL STRUCTURE OF THE COMPLEX OF MICROPLASMINOGEN WITH ALPHA DOMAIN OF STREPTOKINASE IN THE PRESENCE CADMIUM IONS | | Descriptor: | CADMIUM ION, Plasminogen, Streptokinase | | Authors: | Wakeham, N, Terzyan, S, Zhai, P, Loy, J.A, Tang, J, Zhang, X.C. | | Deposit date: | 2002-03-06 | | Release date: | 2002-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Effects of deletion of streptokinase residues 48-59 on plasminogen activation.
PROTEIN ENG., 15, 2002
|
|
1L4D
 
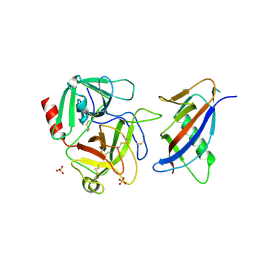 | | CRYSTAL STRUCTURE OF MICROPLASMINOGEN-STREPTOKINASE ALPHA DOMAIN COMPLEX | | Descriptor: | PLASMINOGEN, STREPTOKINASE, SULFATE ION | | Authors: | Wakeham, N, Terzyan, S, Zhai, P, Loy, J.A, Tang, J, Zhang, X.C. | | Deposit date: | 2002-03-04 | | Release date: | 2002-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effects of deletion of streptokinase residues 48-59 on plasminogen activation
PROTEIN ENG., 15, 2002
|
|
1BML
 
 | |
1KTH
 
 | |
6KZF
 
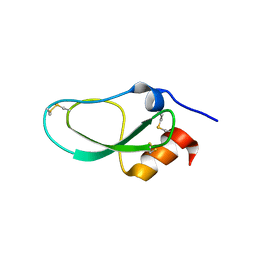 | | Racemic X-ray Structure of Calcicludine | | Descriptor: | D-calcicludine, Kunitz-type serine protease inhibitor homolog calcicludine | | Authors: | Qu, Q, Gao, S, Liu, L. | | Deposit date: | 2019-09-24 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Synthesis of Disulfide Surrogate Peptides Incorporating Large-Span Surrogate Bridges Through a Native-Chemical-Ligation-Assisted Diaminodiacid Strategy
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
3NK4
 
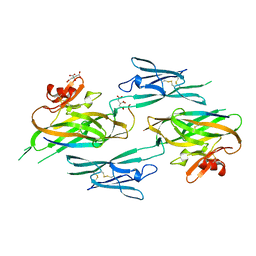 | |
4ES7
 
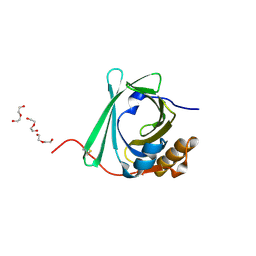 | |
3NAP
 
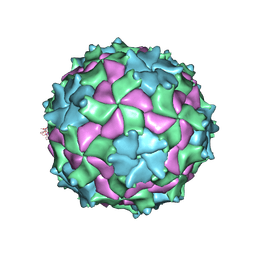 | | Structure of Triatoma Virus (TrV) | | Descriptor: | Capsid protein | | Authors: | Squires, G, Pous, J. | | Deposit date: | 2010-06-02 | | Release date: | 2010-07-28 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystallographic structure of Triatoma Virus (TrV) highlights the Dicistroviridae and Picornaviridae family differences
To be Published
|
|
3NK3
 
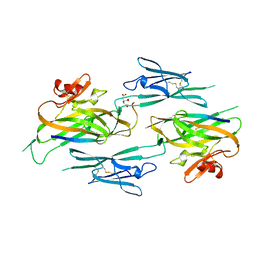 | | Crystal structure of full-length sperm receptor ZP3 at 2.6 A resolution | | Descriptor: | CITRATE ANION, Zona pellucida 3, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Monne, M, Jovine, L. | | Deposit date: | 2010-06-18 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into Egg Coat Assembly and Egg-Sperm Interaction from the X-Ray Structure of Full-Length ZP3.
Cell(Cambridge,Mass.), 143, 2010
|
|
3L4H
 
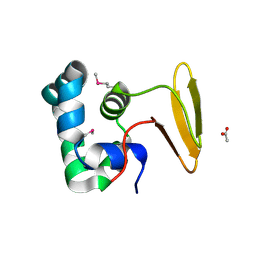 | | Helical box domain and second WW domain of the human E3 ubiquitin-protein ligase HECW1 | | Descriptor: | ACETIC ACID, E3 ubiquitin-protein ligase HECW1 | | Authors: | Walker, J.R, Qiu, L, Li, Y, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Botchkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-12-20 | | Release date: | 2010-05-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The tandem helical box and second WW domains of human HECW1
To be Published
|
|
8CD8
 
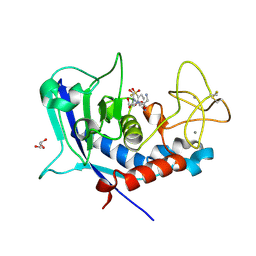 | | Ulilysin - C269A with AEBSF complex | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, CALCIUM ION, GLY-SER-SER, ... | | Authors: | Rodriguez-Banqueri, A, Eckhard, U, Gomis-Ruth, F.X. | | Deposit date: | 2023-01-30 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into latency of the metallopeptidase ulilysin (lysargiNase) and its unexpected inhibition by a sulfonyl-fluoride inhibitor of serine peptidases.
Dalton Trans, 52, 2023
|
|
8CDB
 
 | | Proulilysin E229A structure | | Descriptor: | CALCIUM ION, Ulilysin, ZINC ION | | Authors: | Rodriguez-Banqueri, A, Eckhard, U, Gomis-Ruth, F.X. | | Deposit date: | 2023-01-30 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structural insights into latency of the metallopeptidase ulilysin (lysargiNase) and its unexpected inhibition by a sulfonyl-fluoride inhibitor of serine peptidases.
Dalton Trans, 52, 2023
|
|
3NTG
 
 | | Crystal structure of COX-2 with selective compound 23d-(R) | | Descriptor: | (2R)-6,8-dichloro-7-(2-methylpropoxy)-2-(trifluoromethyl)-2H-chromene-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, J.L, Limburg, D, Graneto, M.J, Carter, J.C, Talley, J.J, Kiefer, J.R. | | Deposit date: | 2010-07-04 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The novel benzopyran class of selective cyclooxygenase-2 inhibitors. Part 2: The second clinical candidate having a shorter and favorable human half-life.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7OXO
 
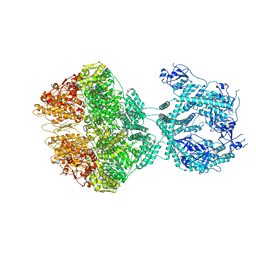 | | human LonP1, R-state, incubated in AMPPCP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Abrahams, J.P, Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T. | | Deposit date: | 2021-06-22 | | Release date: | 2021-12-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
4GPG
 
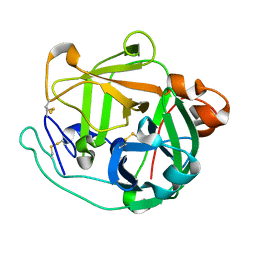 | | X/N joint refinement of Achromobacter Lyticus Protease I free form at pD8.0 | | Descriptor: | Protease 1 | | Authors: | Ohnishi, Y, Yamada, T, Kurihara, K, Tanaka, I, Sakiyama, F, Masaki, T, Niimura, N. | | Deposit date: | 2012-08-21 | | Release date: | 2013-09-11 | | Last modified: | 2024-10-30 | | Method: | NEUTRON DIFFRACTION (1.895 Å), X-RAY DIFFRACTION | | Cite: | Neutron and X-ray crystallographic analysis of Achromobacter protease I at pD 8.0: protonation states and hydration structure in the free-form.
Biochim.Biophys.Acta, 1834, 2013
|
|
2NDN
 
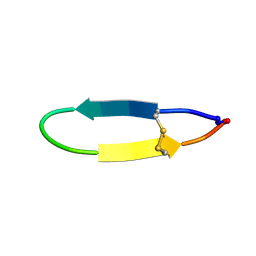 | | NMR solution structure of PawS Derived Peptide 20 (PDP-20) | | Descriptor: | PawS1a Derived Peptide 20 | | Authors: | Franke, B, Jayasena, A.S, Fisher, M.F, Swedberg, J.E, Taylor, N.L, Mylne, J.S, Rosengren, K. | | Deposit date: | 2016-07-26 | | Release date: | 2016-08-24 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Diverse cyclic seed peptides in the Mexican zinnia (Zinnia haageana).
Biopolymers, 106, 2016
|
|
2NDL
 
 | | NMR solution structure of PawS Derived Peptide 22 (PDP-22) | | Descriptor: | PawS derived peptide | | Authors: | Franke, B, Jayasena, A.S, Fisher, M.F, Swedberg, J.E, Taylor, N.L, Mylne, J.S, Rosengren, K. | | Deposit date: | 2016-07-17 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Diverse cyclic seed peptides in the Mexican zinnia (Zinnia haageana).
Biopolymers, 106, 2016
|
|
2NDM
 
 | | NMR solution structure of PawS Derived Peptide 21 (PDP-21) | | Descriptor: | PawS derived peptide 21 | | Authors: | Franke, B, Jayasena, A.S, Fisher, M.F, Swedberg, J.E, Taylor, N.L, Mylne, J.S, Rosengren, K. | | Deposit date: | 2016-07-17 | | Release date: | 2016-08-24 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Diverse cyclic seed peptides in the Mexican zinnia (Zinnia haageana).
Biopolymers, 106, 2016
|
|
1CEH
 
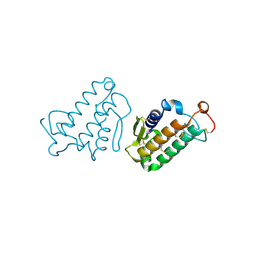 | | STRUCTURE AND FUNCTION OF THE CATALYTIC SITE MUTANT ASP99ASN OF PHOSPHOLIPASE A2: ABSENCE OF CONSERVED STRUCTURAL WATER | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Kumar, A, Sekharudu, C, Ramakrishnan, B, Dupureur, C.M, Zhu, H, Tsai, M.-D, Sundaralingam, M. | | Deposit date: | 1994-11-16 | | Release date: | 1995-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the catalytic site mutant Asp 99 Asn of phospholipase A2: absence of the conserved structural water.
Protein Sci., 3, 1994
|
|
1UEO
 
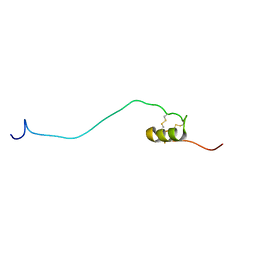 | | Solution structure of the [T8A]-Penaeidin-3 | | Descriptor: | Penaeidin-3a | | Authors: | Yang, Y, Poncet, J, Garnier, J, Zatylny, C, Bachere, E, Aumelas, A. | | Deposit date: | 2003-05-20 | | Release date: | 2003-10-21 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the recombinant penaeidin-3, a shrimp antimicrobial peptide
J.Biol.Chem., 278, 2003
|
|
8HFN
 
 | | Crystal Structure of Mycobacterium smegmatis MshC in Complex with Compound 7b | | Descriptor: | CALCIUM ION, L-cysteine:1D-myo-inositol 2-amino-2-deoxy-alpha-D-glucopyranoside ligase, N-[(3M)-3-(6-methoxypyridin-3-yl)benzene-1-sulfonyl]-L-cysteinamide, ... | | Authors: | Pang, L, Weeks, S.D, Strelkov, S.V. | | Deposit date: | 2022-11-11 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Basis of Cysteine Ligase MshC Inhibition by Cysteinyl-Sulfonamides.
Int J Mol Sci, 23, 2022
|
|
8HFM
 
 | | Crystal Structure of Mycobacterium smegmatis MshC | | Descriptor: | CALCIUM ION, L-cysteine:1D-myo-inositol 2-amino-2-deoxy-alpha-D-glucopyranoside ligase, ZINC ION | | Authors: | Pang, L, Weeks, S.D, Strelkov, S.V. | | Deposit date: | 2022-11-11 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural Basis of Cysteine Ligase MshC Inhibition by Cysteinyl-Sulfonamides.
Int J Mol Sci, 23, 2022
|
|
8HFO
 
 | | Crystal Structure of Mycobacterium smegmatis MshC in Complex with Compound 7d | | Descriptor: | CALCIUM ION, L-cysteine:1D-myo-inositol 2-amino-2-deoxy-alpha-D-glucopyranoside ligase, N-[(3M)-3-(thiophen-2-yl)benzene-1-sulfonyl]-L-cysteinamide, ... | | Authors: | Pang, L, Weeks, S.D, Strelkov, S.V. | | Deposit date: | 2022-11-11 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural Basis of Cysteine Ligase MshC Inhibition by Cysteinyl-Sulfonamides.
Int J Mol Sci, 23, 2022
|
|
8HU7
 
 | |
