2B0K
 
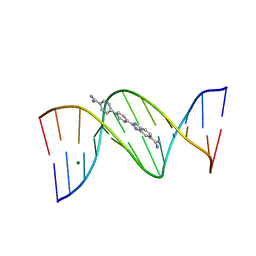 | | Crystal structure of the DB921-D(CGCGAATTCGCG)2 complex. | | Descriptor: | 2-(4'-AMIDINOBIPHENYL-4-YL)-1H-BENZIMIDAZOLE-5-AMIDINE, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION | | Authors: | Miao, Y, Lee, M.P, Parkinson, G.N, Batista-Parra, A, Ismail, M.A, Neidle, S, Boykin, D.W, Wilson, W.D. | | Deposit date: | 2005-09-14 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Out-of-Shape DNA Minor Groove Binders: Induced Fit Interactions of Heterocyclic Dications with the DNA Minor Groove.
Biochemistry, 44, 2005
|
|
2B0L
 
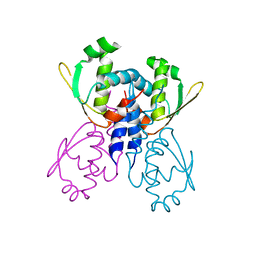 | | C-terminal DNA binding domain of transcriptional pleiotropic repressor CodY. | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor codY | | Authors: | Levdikov, V.M, Blagova, E, Joseph, P, Sonenshein, A.L, Wilkinson, A.J. | | Deposit date: | 2005-09-14 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of CodY, a GTP- and Isoleucine-responsive Regulator of Stationary Phase and Virulence in Gram-positive Bacteria.
J.Biol.Chem., 281, 2006
|
|
2B0M
 
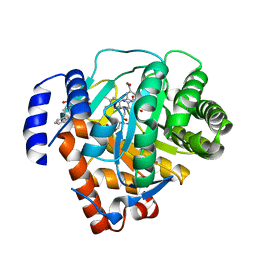 | | Human dihydroorotate dehydrogenase bound to a novel inhibitor | | Descriptor: | 3-AMIDO-5-BIPHENYL-BENZOIC ACID, Dihydroorotate dehydrogenase, mitochondrial, ... | | Authors: | Hurt, D.E, Sutton, A.E, Clardy, J. | | Deposit date: | 2005-09-14 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Brequinar derivatives and species-specific drug design for dihydroorotate dehydrogenase.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2B0O
 
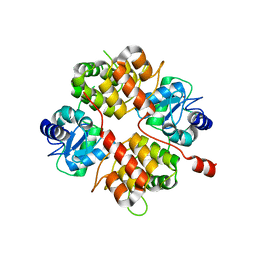 | | Crystal structure of UPLC1 GAP domain | | Descriptor: | UPLC1, ZINC ION | | Authors: | Ismail, S, Shen, L, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-09-14 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural analysis of GAP and ankyrin domains of UPLC1
To be Published
|
|
2B0P
 
 | | truncated S. aureus LytM, P212121 crystal form | | Descriptor: | ACETATE ION, CACODYLATE ION, Glycyl-glycine endopeptidase lytM, ... | | Authors: | Firczuk, M, Mucha, A, Bochtler, M. | | Deposit date: | 2005-09-14 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of active LytM.
J.Mol.Biol., 354, 2005
|
|
2B0Q
 
 | |
2B0R
 
 | | Crystal Structure of Cyclase-Associated Protein from Cryptosporidium parvum | | Descriptor: | UNKNOWN ATOM OR ION, possible adenyl cyclase-associated protein | | Authors: | Tempel, W, Dong, A, Zhao, Y, Lew, J, Kozieradzki, I, Alam, Z, Melone, M, Wasney, G, Vedadi, M, Arrowsmith, C, Edwards, A, Weigelt, J, Sundstrom, M, Hui, R, Bochkarev, A, Artz, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-09-14 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and function of a G-actin sequestering protein with a vital role in malaria oocyst development inside the mosquito vector
J.Biol.Chem., 151, 2010
|
|
2B0S
 
 | | Crystal structure analysis of anti-HIV-1 V3 Fab 2219 in complex with MN peptide | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Fab 2219, ... | | Authors: | Stanfield, R.L, Gorny, M.K, Zolla-Pazner, S, Wilson, I.A. | | Deposit date: | 2005-09-14 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of human immunodeficiency virus type 1 (HIV-1) neutralizing antibody 2219 in complex with three different V3 peptides reveal a new binding mode for HIV-1 cross-reactivity.
J.Virol., 80, 2006
|
|
2B0T
 
 | | Structure of Monomeric NADP Isocitrate dehydrogenase | | Descriptor: | MAGNESIUM ION, NADP Isocitrate dehydrogenase | | Authors: | Imabayashi, F, Aich, S, Prasad, L, Delbaere, L.T. | | Deposit date: | 2005-09-14 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Substrate-free structure of a monomeric NADP isocitrate dehydrogenase: An open conformation phylogenetic relationship of isocitrate dehydrogenase.
Proteins, 63, 2006
|
|
2B0U
 
 | | The Structure of the Follistatin:Activin Complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Follistatin, IRIDIUM (III) ION, ... | | Authors: | Thompson, T.B, Lerch, T.F, Cook, R.W, Woodruff, T.K, Jardetzky, T.S. | | Deposit date: | 2005-09-14 | | Release date: | 2005-10-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of the Follistatin:Activin Complex Reveals Antagonism of Both Type I and Type II Receptor Binding.
Dev.Cell, 9, 2005
|
|
2B0V
 
 | | NUDIX hydrolase from Nitrosomonas europaea. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, NUDIX hydrolase, ... | | Authors: | Osipiuk, J, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-09-14 | | Release date: | 2005-09-27 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray structure of NUDIX hydrolase from Nitrosomonas europaea.
To be Published
|
|
2B0Y
 
 | | Solution Structure of a peptide mimetic of the fourth cytoplasmic loop of the G-protein coupled CB1 cannabinoid receptor | | Descriptor: | Cannabinoid receptor 1 | | Authors: | Grace, C.R, Cowsik, S.M, Shim, J.Y, Welsh, W.J, Howlett, A.C. | | Deposit date: | 2005-09-15 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Unique helical conformation of the fourth cytoplasmic loop of the CB1 cannabinoid receptor in a negatively charged environment.
J.Struct.Biol., 159, 2007
|
|
2B0Z
 
 | |
2B10
 
 | |
2B11
 
 | |
2B12
 
 | |
2B13
 
 | | Truncated S. aureus LytM, P41 crystal form | | Descriptor: | Glycyl-glycine endopeptidase lytM, L(+)-TARTARIC ACID, ZINC ION | | Authors: | Firczuk, M, Mucha, A, Bochtler, M. | | Deposit date: | 2005-09-15 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of active LytM.
J.Mol.Biol., 354, 2005
|
|
2B14
 
 | | The crystal structure of 2,4-dinitrophenol in complex with the amyloidogenic variant Transthyretin Leu 55 Pro | | Descriptor: | 2,4-DINITROPHENOL, Transthyretin | | Authors: | Morais-de-Sa, E, Neto-Silva, R.M, Pereira, P.J, Saraiva, M.J, Damas, A.M. | | Deposit date: | 2005-09-15 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The binding of 2,4-dinitrophenol to wild-type and amyloidogenic transthyretin
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2B15
 
 | | The crystal structure of 2,4-dinitrophenol in complex with human transthyretin | | Descriptor: | 2,4-DINITROPHENOL, SULFATE ION, Transthyretin | | Authors: | Morais-de-Sa, E, Neto-Silva, R.M, Pereira, P.J, Saraiva, M.J, Damas, A.M. | | Deposit date: | 2005-09-15 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The binding of 2,4-dinitrophenol to wild-type and amyloidogenic transthyretin
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2B16
 
 | | The crystal structure of 2,4-dinitrophenol in complex with the amyloidogenic variant Transthyretin Tyr78Phe | | Descriptor: | 2,4-DINITROPHENOL, Transthyretin | | Authors: | Morais-de-Sa, E, Neto-Silva, R.M, Pereira, P.J, Saraiva, M.J, Damas, A.M. | | Deposit date: | 2005-09-15 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The binding of 2,4-dinitrophenol to wild-type and amyloidogenic transthyretin
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2B17
 
 | | Specific binding of non-steroidal anti-inflammatory drugs (NSAIDs) to phospholipase A2: Crystal structure of the complex formed between phospholipase A2 and diclofenac at 2.7 A resolution: | | Descriptor: | 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Singh, N, Jabeen, T, Sharma, S, Singh, T.P. | | Deposit date: | 2005-09-15 | | Release date: | 2005-10-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Specific binding of non-steroidal anti-inflammatory drugs (NSAIDs) to phospholipase A2: structure of the complex formed between phospholipase A2 and diclofenac at 2.7 A resolution.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2B18
 
 | | N-terminal GAF domain of transcriptional pleiotropic repressor CodY. | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor codY, ISOLEUCINE | | Authors: | Levdikov, V.M, Blagova, E, Joseph, P, Sonenshein, A.L, Wilkinson, A.J. | | Deposit date: | 2005-09-15 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of CodY, a GTP- and Isoleucine-responsive Regulator of Stationary Phase and Virulence in Gram-positive Bacteria.
J.Biol.Chem., 281, 2006
|
|
2B19
 
 | |
2B1A
 
 | | Crystal structure analysis of anti-HIV-1 V3 Fab 2219 in complex with UG1033 peptide | | Descriptor: | Fab 2219, heavy chain, light chain, ... | | Authors: | Stanfield, R.L, Gorny, M.K, Zolla-Pazner, S, Wilson, I.A. | | Deposit date: | 2005-09-15 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Crystal structures of human immunodeficiency virus type 1 (HIV-1) neutralizing antibody 2219 in complex with three different V3 peptides reveal a new binding mode for HIV-1 cross-reactivity.
J.Virol., 80, 2006
|
|
2B1B
 
 | |
