2AP9
 
 | | Crystal structure of acetylglutamate kinase from Mycobacterium tuberculosis CDC1551 | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, acetylglutamate kinase | | Authors: | Rajashankar, K.R, Kniewel, R, Lee, K, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-08-15 | | Release date: | 2005-08-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of acetylglutamate kinase from Mycobacterium tuberculosis CDC1551
To be Published
|
|
2APB
 
 | | Crystal Structure of the S54N variant of murine T cell receptor Vbeta 8.2 domain | | Descriptor: | MALONIC ACID, T-cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2APC
 
 | |
2APF
 
 | | Crystal Structure of the A52V/S54N/K66E variant of the murine T cell receptor V beta 8.2 domain | | Descriptor: | MALONIC ACID, T cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2APG
 
 | | The structure of tryptophan 7-halogenase (PrnA)suggests a mechanism for regioselective chlorination | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Dong, C, Flecks, S, Unversucht, S, Haupt, C, Van Pee, K.H, Naismith, J.H, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2005-08-16 | | Release date: | 2005-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tryptophan 7-halogenase (PrnA) structure suggests a mechanism for regioselective chlorination.
Science, 309, 2005
|
|
2APH
 
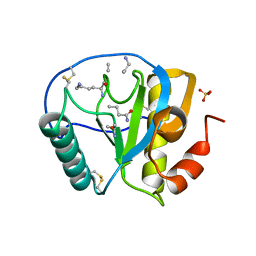 | | Crystal structure of human PGRP-IalphaC in complex with muramyl pentapeptide | | Descriptor: | N-acetyl-beta-muramic acid, Peptidoglycan recognition protein I-alpha, SULFATE ION, ... | | Authors: | Guan, R, Roychowdjury, A, Boons, G, Mariuzza, R.A. | | Deposit date: | 2005-08-16 | | Release date: | 2006-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human peptidoglycan recognition protein I alpha bound to a muramyl pentapeptide from Gram-positive bacteria.
Protein Sci., 15, 2006
|
|
2APJ
 
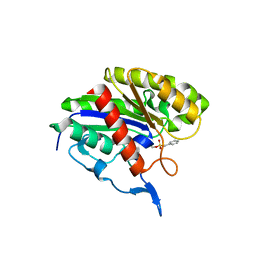 | | X-Ray Structure of Protein from Arabidopsis Thaliana AT4G34215 at 1.6 Angstrom Resolution | | Descriptor: | Putative Esterase | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bitto, E, Bingman, C.A, Allard, S.T, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-08-16 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure at 1.6 Angstroms resolution of the protein product of the At4g34215 gene from Arabidopsis thaliana.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2APL
 
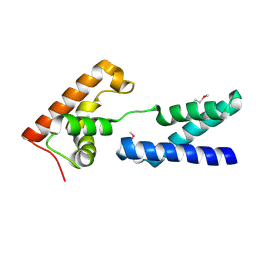 | | Crystal structure of protein PG0816 from Porphyromonas gingivalis | | Descriptor: | hypothetical protein PG0816 | | Authors: | Chang, C, Quartey, P, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-16 | | Release date: | 2005-09-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of protein PG0816 from Porphyromonas gingivalis
To be Published
|
|
2APN
 
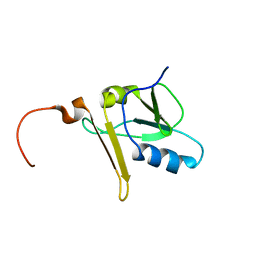 | |
2APO
 
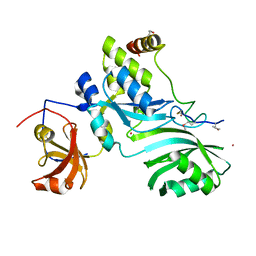 | | Crystal Structure of the Methanococcus jannaschii Cbf5 Nop10 Complex | | Descriptor: | POTASSIUM ION, Probable tRNA pseudouridine synthase B, Ribosome biogenesis protein Nop10, ... | | Authors: | Hamma, T, Reichow, S.L, Varani, G, Ferre-D'Amare, A.R. | | Deposit date: | 2005-08-16 | | Release date: | 2005-11-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Cbf5-Nop10 complex is a molecular bracket that organizes box H/ACA RNPs.
Nat.Struct.Mol.Biol., 12, 2005
|
|
2APQ
 
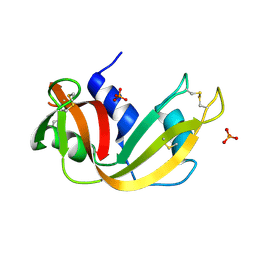 | | Crystal Structure of an Active Site Mutant of Bovine Pancreatic Ribonuclease A (H119A-RNase A) with a 10-Glutamine expansion in the C-terminal hinge-loop. | | Descriptor: | PHOSPHATE ION, Ribonuclease | | Authors: | Sambashivan, S, Liu, Y, Sawaya, M.R, Gingery, M, Eisenberg, D. | | Deposit date: | 2005-08-16 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Amyloid-like fibrils of ribonuclease A with three-dimensional domain-swapped and native-like structure.
Nature, 437, 2005
|
|
2APR
 
 | |
2APS
 
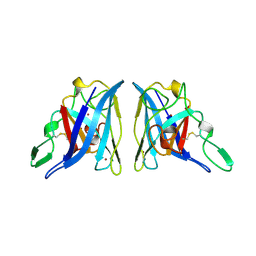 | | CU/ZN SUPEROXIDE DISMUTASE FROM ACTINOBACILLUS PLEUROPNEUMONIAE | | Descriptor: | COPPER (II) ION, PROTEIN (CU,ZN SUPEROXIDE DISMUTASE), ZINC ION | | Authors: | Forest, K.T, Langford, P.R, Kroll, J.S, Getzoff, E.D. | | Deposit date: | 1999-02-11 | | Release date: | 1999-02-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cu,Zn superoxide dismutase structure from a microbial pathogen establishes a class with a conserved dimer interface.
J.Mol.Biol., 296, 2000
|
|
2APT
 
 | | Crystal Structure of the G17E/S54N/K66E/Q72H/E80V/L81S/T87S/G96V variant of the murine T cell receptor V beta 8.2 domain | | Descriptor: | MALONIC ACID, T-cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2APV
 
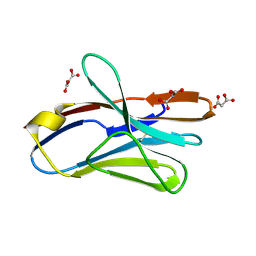 | | Crystal Structure of the G17E/A52V/S54N/Q72H/E80V/L81S/T87S/G96V variant of the murine T cell receptor V beta 8.2 domain | | Descriptor: | MALONIC ACID, T cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2APW
 
 | | Crystal Structure of the G17E/A52V/S54N/K66E/E80V/L81S/T87S/G96V variant of the murine T cell receptor V beta 8.2 domain | | Descriptor: | MALONIC ACID, T cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2APX
 
 | | Crystal Structure of the G17E/A52V/S54N/K66E/Q72H/E80V/L81S/T87S/G96V variant of the murine T cell receptor V beta 8.2 domain | | Descriptor: | MALONIC ACID, T cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2AQ0
 
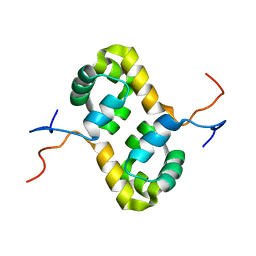 | | Solution structure of the human homodimeric dna repair protein XPF | | Descriptor: | DNA repair endonuclease XPF | | Authors: | Das, D, Tripsianes, K, Folkers, G, Jaspers, N.G, Hoeijmakers, J.H, Kaptein, R, Boelens, R. | | Deposit date: | 2005-08-17 | | Release date: | 2006-10-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The HhH domain of the human DNA repair protein XPF forms stable homodimers
Proteins, 70, 2008
|
|
2AQ1
 
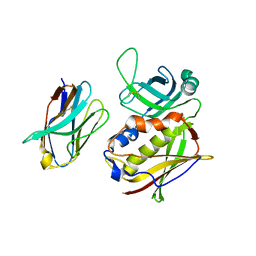 | | Crystal structure of T-cell receptor V beta domain variant complexed with superantigen SEC3 mutant | | Descriptor: | Enterotoxin type C-3, T-cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-17 | | Release date: | 2006-03-21 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2AQ2
 
 | | Crystal structure of T-cell receptor V beta domain variant complexed with superantigen SEC3 mutant | | Descriptor: | Enterotoxin type C-3, SODIUM ION, SULFATE ION, ... | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-17 | | Release date: | 2006-03-21 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2AQ3
 
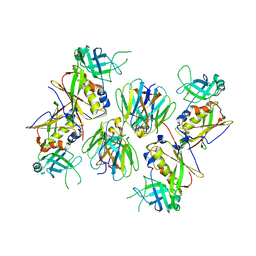 | | Crystal structure of T-cell receptor V beta domain variant complexed with superantigen SEC3 | | Descriptor: | Enterotoxin type C-3, T-cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-17 | | Release date: | 2006-03-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2AQ4
 
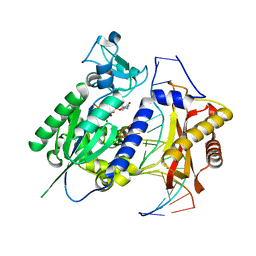 | | Ternary complex of the catalytic core of REV1 with DNA and dCTP. | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*AP*TP*CP*CP*TP*CP*CP*CP*CP*TP*AP*C)-3', 5'-D(*TP*AP*AP*GP*GP*TP*AP*GP*GP*GP*GP*AP*GP*GP*AP*T)-3', ... | | Authors: | Nair, D.T, Johnson, R.E, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2005-08-17 | | Release date: | 2005-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Rev1 employs a novel mechanism of DNA synthesis using a protein template.
Science, 309, 2005
|
|
2AQ5
 
 | |
2AQ6
 
 | | X-ray crystal structure of mycobacterium tuberculosis pyridoxine 5'-phosphate oxidase complexed with pyridoxal 5'-phosphate at 1.7 a resolution | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, PYRIDOXINE 5'-PHOSPHATE OXIDASE | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Garen, C, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-08-17 | | Release date: | 2005-08-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of Mycobacterium tuberculosispyridoxine 5'-phosphate oxidase and its complexes with flavin mononucleotide and pyridoxal 5'-phosphate.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2AQ7
 
 | | Structure-activity relationships at the 5-posiiton of thiolactomycin: an intact 5(R)-isoprene unit is required for activity against the condensing enzymes from Mycobacterium tuberculosis and Escherichia coli | | Descriptor: | (5R)-4-HYDROXY-3,5-DIMETHYL-5-[(1E,3E)-2-METHYLPENTA-1,3-DIENYL]THIOPHEN-2(5H)-ONE, 3-oxoacyl-[acyl-carrier-protein] synthase I | | Authors: | Kim, P, Zhang, Y.M, Shenoy, G, Nguyen, Q.A, Boshoff, H.I, Manjunatha, U.H, Goodwin, M.B, Lonsdale, J, Price, A.C, Miller, D.J. | | Deposit date: | 2005-08-17 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Activity Relationships at the 5-Position of Thiolactomycin: An Intact (5R)-Isoprene Unit Is Required for Activity against the Condensing Enzymes from Mycobacterium tuberculosis and Escherichia coli
J.Med.Chem., 49, 2006
|
|
