3V9R
 
 | | Crystal structure of Saccharomyces cerevisiae MHF complex | | Descriptor: | SULFATE ION, Uncharacterized protein YDL160C-A, Uncharacterized protein YOL086W-A | | Authors: | Yang, H, Zhang, T, Zhong, C, Li, H, Zhou, J, Ding, J. | | Deposit date: | 2011-12-28 | | Release date: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Saccharomyces Cerevisiae MHF Complex Structurally Resembles the Histones (H3-H4)(2) Heterotetramer and Functions as a Heterotetramer
Structure, 20, 2012
|
|
2J3T
 
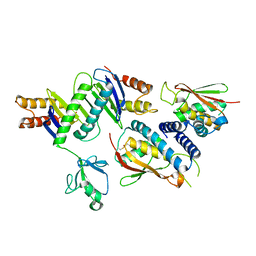 | | The crystal structure of the bet3-trs33-bet5-trs23 complex. | | Descriptor: | PALMITIC ACID, TRAFFICKING PROTEIN PARTICLE COMPLEX SUBUNIT 1, TRAFFICKING PROTEIN PARTICLE COMPLEX SUBUNIT 3, ... | | Authors: | Kim, Y, Oh, B. | | Deposit date: | 2006-08-23 | | Release date: | 2006-11-22 | | Last modified: | 2011-10-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Architecture of the Multisubunit Trapp I Complex Suggests a Model for Vesicle Tethering.
Cell(Cambridge,Mass.), 127, 2006
|
|
1NYD
 
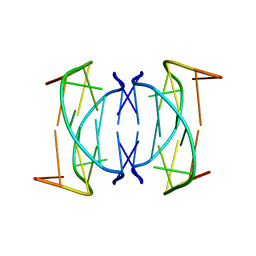 | | Solution structure of DNA quadruplex GCGGTGGAT | | Descriptor: | 5'-D(*GP*CP*GP*GP*TP*GP*GP*AP*T)-3' | | Authors: | Webba da Silva, M. | | Deposit date: | 2003-02-12 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Association of DNA quadruplexes through G:C:G:C tetrads. Solution structure of d(GCGGTGGAT).
Biochemistry, 42, 2003
|
|
7WU5
 
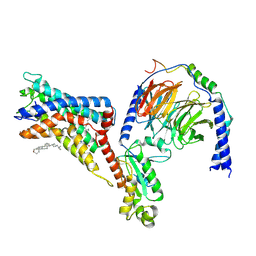 | | Cryo-EM structure of the adhesion GPCR ADGRF1(H565A/T567A) in complex with miniGi | | Descriptor: | Adhesion G-protein coupled receptor F1, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2022-02-05 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
7WU4
 
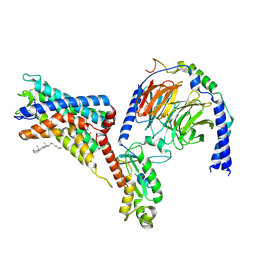 | | Cryo-EM structure of the adhesion GPCR ADGRF1 in complex with miniGi | | Descriptor: | Adhesion G-protein coupled receptor F1, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2022-02-05 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
3VD7
 
 | | E. coli (lacZ) beta-galactosidase (N460S) in complex with galactotetrazole | | Descriptor: | (5R, 6S, 7S, ... | | Authors: | Wheatley, R.W, Kappelhoff, J.C, Hahn, J.N, Dugdale, M.L, Dutkoski, M.J, Tamman, S.D, Fraser, M.E, Huber, R.E. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Substitution for Asn460 cripples {beta}-galactosidase (Escherichia coli) by increasing substrate affinity and decreasing transition state stability.
Arch.Biochem.Biophys., 521, 2012
|
|
1Y76
 
 | | Solution Structure of Patj/Pals1 L27 Domain Complex | | Descriptor: | MAGUK p55 subfamily member 5, protein associated to tight junctions | | Authors: | Feng, W, Long, J.-F, Zhang, M. | | Deposit date: | 2004-12-08 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A unified assembly mode revealed by the structures of tetrameric L27 domain complexes formed by mLin-2/mLin-7 and Patj/Pals1 scaffold proteins.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1Y74
 
 | | Solution Structure of mLin-2/mLin-7 L27 Domain Complex | | Descriptor: | Peripheral plasma membrane protein CASK, lin 7 homolog b | | Authors: | Feng, W, Long, J.-F, Zhang, M. | | Deposit date: | 2004-12-08 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A unified assembly mode revealed by the structures of tetrameric L27 domain complexes formed by mLin-2/mLin-7 and Patj/Pals1 scaffold proteins.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1V3O
 
 | | Crystal structure of d(GCGAGAGC): the DNA quadruplex structure split from the octaplex | | Descriptor: | 5'-D(*GP*(C38)P*GP*AP*GP*AP*GP*C)-3', POTASSIUM ION | | Authors: | Kondo, J, Umeda, S, Sunami, T, Takenaka, A. | | Deposit date: | 2003-11-03 | | Release date: | 2004-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of a DNA octaplex with I-motif of G-quartets and its splitting into two quadruplexes suggest a folding mechanism of eight tandem repeats
Nucleic Acids Res., 32, 2004
|
|
1V3N
 
 | | Crystal structure of d(GCGAGAGC): the DNA quadruplex structure split from the octaplex | | Descriptor: | 5'-D(*GP*(CBR)P*GP*AP*GP*AP*GP*C)-3', POTASSIUM ION | | Authors: | Kondo, J, Umeda, S, Sunami, T, Takenaka, A. | | Deposit date: | 2003-11-03 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of a DNA octaplex with I-motif of G-quartets and its splitting into two quadruplexes suggest a folding mechanism of eight tandem repeats
Nucleic Acids Res., 32, 2004
|
|
1QX8
 
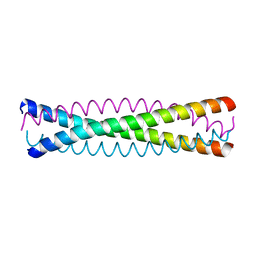 | | Crystal structure of a five-residue deletion mutant of the Rop protein | | Descriptor: | Regulatory protein ROP | | Authors: | Glykos, N.M, Vlassi, M, Papanikolaou, Y, Kotsifaki, D, Cesareni, G, Kokkinidis, M. | | Deposit date: | 2003-09-04 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Loopless Rop: structure and dynamics of an engineered homotetrameric variant of the repressor of primer protein.
Biochemistry, 45, 2006
|
|
6X4N
 
 | | Human cyclophilin A bound to a series of acylcic and macrocyclic inhibitors: (2R,5S,11S,14S,18E)-2,11,17,17-tetramethyl-14-(propan-2-yl)-3-oxa-9,12,15,26,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(25),18,20(28),21,23,26-hexaene-4,10,13,16-tetrone (compound 24) | | Descriptor: | (2R,5S,11S,14S,18E)-2,11,17,17-tetramethyl-14-(propan-2-yl)-3-oxa-9,12,15,26,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(25),18,20(28),21,23,26-hexaene-4,10,13,16-tetrone, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Appleby, T.C, Paulsen, J.L, Schmitz, U, Shivakumar, D. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Evaluation of Free Energy Calculations for the Prioritization of Macrocycle Synthesis.
J.Chem.Inf.Model., 60, 2020
|
|
6X4P
 
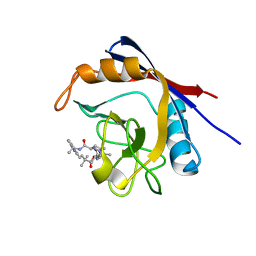 | | Human cyclophilin A bound to a series of acylcic and macrocyclic inhibitors: (2R,5S,11S,14S,18E)-2,11,17,17-tetramethyl-14-(propan-2-yl)-15-oxa-3,9,12,26,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(25),18,20(28),21,23,26-hexaene-4,10,13,16-tetrone (compound 28) | | Descriptor: | (2R,5S,11S,14S,18E)-2,11,17,17-tetramethyl-14-(propan-2-yl)-15-oxa-3,9,12,26,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(25),18,20(28),21,23,26-hexaene-4,10,13,16-tetrone, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Appleby, T.C, Paulsen, J.L, Schmitz, U, Shivakumar, D. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Evaluation of Free Energy Calculations for the Prioritization of Macrocycle Synthesis.
J.Chem.Inf.Model., 60, 2020
|
|
6X4Q
 
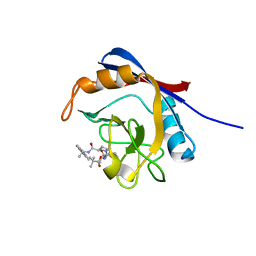 | | Human cyclophilin A bound to a series of acylcic and macrocyclic inhibitors: (2R,5S,11S,14S,18E)-14-cyclobutyl-2,11,17,17-tetramethyl-15-oxa-3,9,12,26,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(25),18,20(28),21,23,26-hexaene-4,10,13,16-tetrone (compound 33) | | Descriptor: | (2R,5S,11S,14S,18E)-14-cyclobutyl-2,11,17,17-tetramethyl-15-oxa-3,9,12,26,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(25),18,20(28),21,23,26-hexaene-4,10,13,16-tetrone, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Appleby, T.C, Paulsen, J.L, Schmitz, U, Shivakumar, D. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of Free Energy Calculations for the Prioritization of Macrocycle Synthesis.
J.Chem.Inf.Model., 60, 2020
|
|
1RWO
 
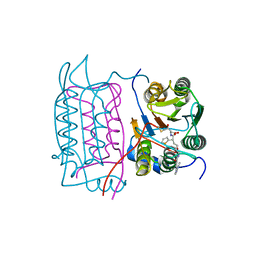 | | Crystal structure of human caspase-1 in complex with 4-oxo-3-{6-[4-(quinoxalin-2-ylamino)-benzoylamino]-2-thiophen-2-yl-hexanoylamino}-pentanoic acid | | Descriptor: | 4-OXO-3-{6-[4-(QUINOXALIN-2-YLAMINO)-BENZOYLAMINO]-2-THIOPHEN-2-YL-HEXANOYLAMINO}-PENTANOIC ACID, Interleukin-1 beta convertase | | Authors: | Romanowski, M.J, Waal, N.D, Fahr, B.T, O'Brien, T. | | Deposit date: | 2003-12-16 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of caspase-1 inhibitors derived from Tethering.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1RWN
 
 | |
1RWM
 
 | | Crystal structure of human caspase-1 in complex with 4-oxo-3-[2-(5-{[4-(quinoxalin-2-ylamino)-benzoylamino]-methyl}-thiophen-2-yl)-acetylamino]-pentanoic acid | | Descriptor: | 4-OXO-3-[2-(5-{[4-(QUINOXALIN-2-YLAMINO)-BENZOYLAMINO]-METHYL}-THIOPHEN-2-YL)-ACETYLAMINO]-PENTANOIC ACID, Interleukin-1 beta convertase | | Authors: | Romanowski, M.J, Waal, N.D, Fahr, B.T, O'Brien, T. | | Deposit date: | 2003-12-16 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of caspase-1 inhibitors derived from Tethering.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1RWP
 
 | |
1RWK
 
 | | Crystal structure of human caspase-1 in complex with 3-(2-mercapto-acetylamino)-4-oxo-pentanoic acid | | Descriptor: | 3-(2-MERCAPTO-ACETYLAMINO)-4-OXO-PENTANOIC ACID, Interleukin-1 beta convertase | | Authors: | Romanowski, M.J, Lam, J.W, Fahr, B.T, O'Brien, T. | | Deposit date: | 2003-12-16 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of caspase-1 inhibitors derived from Tethering.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1XOE
 
 | | N9 Tern influenza neuraminidase complexed with (2R,4R,5R)-5-(1-Acetylamino-3-methyl-butyl-pyrrolidine-2, 4-dicarobyxylic acid 4-methyl esterdase complexed with | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[1-(ACETYLAMINO)-3-METHYLBUTYL]-4-(METHOXYCARBONYL)PROLINE, Neuraminidase, ... | | Authors: | Wang, G.T, Wang, S, Gentles, R, Sowin, T, Maring, C.J, Kempf, D.J, Kati, W.M, Stoll, V, Stewart, K.D, Laver, G. | | Deposit date: | 2004-10-06 | | Release date: | 2005-01-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, synthesis, and structural analysis of inhibitors of influenza neuraminidase
containing a 2,3-disubstituted tetrahydrofuran-5-carboxylic acid core.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1VAR
 
 | |
1OU6
 
 | |
1TT0
 
 | | Crystal Structure of Pyranose 2-Oxidase | | Descriptor: | ACETATE ION, DODECAETHYLENE GLYCOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hallberg, B.M, Leitner, C, Haltrich, D, Divne, C. | | Deposit date: | 2004-06-21 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the 270 kDa homotetrameric lignin-degrading enzyme pyranose 2-oxidase
J.Mol.Biol., 341, 2004
|
|
1SN0
 
 | | Crystal Structure Of Sea Bream Transthyretin in complex with thyroxine At 1.9A Resolution | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, SULFATE ION, transthyretin | | Authors: | Eneqvist, T, Lundberg, E, Karlsson, A, Huang, S, Santos, C.R, Power, D.M, Sauer-Eriksson, A.E. | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High resolution crystal structures of piscine transthyretin reveal different binding modes for triiodothyronine and thyroxine.
J.Biol.Chem., 279, 2004
|
|
1SQL
 
 | | Crystal structure of 7,8-dihydroneopterin aldolase in complex with guanine | | Descriptor: | GUANINE, dihydroneopterin aldolase | | Authors: | Bauer, S, Schott, A.K, Illarionova, V, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2004-03-19 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biosynthesis of Tetrahydrofolate in Plants: Crystal Structure of 7,8-Dihydroneopterin Aldolase from Arabidopsis thaliana Reveals a Novel Adolase Class.
J.Mol.Biol., 339, 2004
|
|
