8P67
 
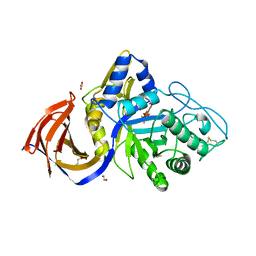 | | Crystal structure of Thermothelomyces thermophila (double mutant EE) in complex with aldotetrauronic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, GH30 family xylanase, ... | | Authors: | Dimarogona, M, Pentari, C, Kosinas, C, Topakas, E. | | Deposit date: | 2023-05-25 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural and molecular insights into a bifunctional glycoside hydrolase 30 xylanase specific to glucuronoxylan.
Biotechnol.Bioeng., 121, 2024
|
|
7PHF
 
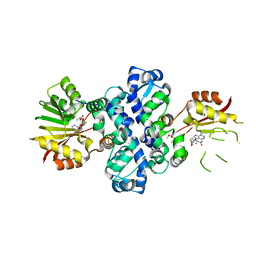 | | Chimeric carminomycin-4-O-methyltransferase (DnrK) with regions from 10-hydroxylase RdmB and 10-decarboxylase TamK | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein,Aclacinomycin 10-hydroxylase RdmB, S-ADENOSYL-L-HOMOCYSTEINE, methyl (1R,2R,4S)-2-ethyl-2,4,5,7-tetrahydroxy-6,11-dioxo-1,2,3,4,6,11-hexahydrotetracene-1-carboxylate | | Authors: | Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-08-17 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
7AO9
 
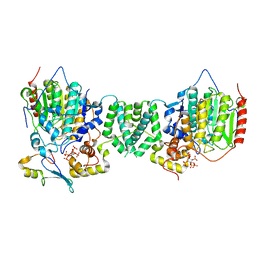 | | Structure of the core MTA1/HDAC1/MBD2 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, Metastasis-associated protein MTA1, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
7E8H
 
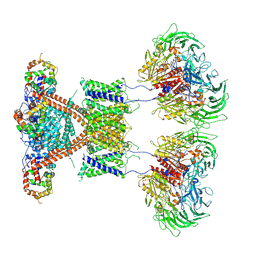 | | CryoEM structure of human Kv4.2-DPP6S-KChIP1 complex | | Descriptor: | Dipeptidyl aminopeptidase-like protein 6, Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Kise, Y, Nureki, O. | | Deposit date: | 2021-03-01 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of gating modulation of Kv4 channel complexes.
Nature, 599, 2021
|
|
7PXR
 
 | | Room temperature structure of an LPMO. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, CHLORIDE ION, ... | | Authors: | Tandrup, T, Meilleur, F, Ipsen, J, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
8P14
 
 | | USP28 USP domain in complex with Vismodegib | | Descriptor: | 2-chloranyl-~{N}-(4-chloranyl-3-pyridin-2-yl-phenyl)-4-methylsulfonyl-benzamide, CHLORIDE ION, Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Sauer, F, Karal Nair, R, Kisker, C. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | USP28 USP domain in complex with Vismodegib
To Be Published
|
|
5L7P
 
 | | In silico-powered specific incorporation of photocaged Dopa at multiple protein sites | | Descriptor: | (2~{S})-2-azanyl-3-[3-[(2-nitrophenyl)methoxy]-4-oxidanyl-phenyl]propanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hauf, M, Richter, F, Schneider, T, Martins, B.M, Baumann, T, Durkin, P, Dobbek, H, Moeglich, A, Budisa, N. | | Deposit date: | 2016-06-03 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Photoactivatable Mussel-Based Underwater Adhesive Proteins by an Expanded Genetic Code.
Chembiochem, 18, 2017
|
|
2ZFF
 
 | | Exploring Thrombin S1-pocket | | Descriptor: | D-phenylalanyl-N-benzyl-L-prolinamide, Hirudin variant-1, SODIUM ION, ... | | Authors: | Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-01-04 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Exploring Thrombin S1-pocket
To be Published
|
|
2M8X
 
 | | Restrained CS-Rosetta Solution NMR structure of the CARDB domain of PF1109 from Pyrococcus furiosus. Northeast Structural Genomics Consortium target PfR193A | | Descriptor: | Uncharacterized protein | | Authors: | Mao, B, Tejero, R.T, Aramini, J.M, Snyder, D.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | PDBStat: a universal restraint converter and restraint analysis software package for protein NMR.
J.Biomol.Nmr, 56, 2013
|
|
7PXJ
 
 | | X-ray structure of LPMO at 5.99x10^4 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, COPPER (II) ION | | Authors: | Tandrup, T, Lo Leggio, L. | | Deposit date: | 2021-10-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
8P1P
 
 | | USP28 in complex with AZ1 | | Descriptor: | 2-[[5-bromanyl-2-[[4-fluoranyl-3-(trifluoromethyl)phenyl]methoxy]phenyl]methylamino]ethanol, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Sauer, F, Karal-Nair, R, Kisker, C. | | Deposit date: | 2023-05-12 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | USP28 in complex with AZ1
To Be Published
|
|
5L0F
 
 | |
2M56
 
 | | The structure of the complex of cytochrome P450cam and its electron donor putidaredoxin determined by paramagnetic NMR spectroscopy | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Hiruma, Y, Hass, M.A.S, Ubbink, M. | | Deposit date: | 2013-02-14 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of the cytochrome p450cam-putidaredoxin complex determined by paramagnetic NMR spectroscopy and crystallography.
J.Mol.Biol., 425, 2013
|
|
7AO0
 
 | | Crystal structure of CotB2 variant F107A in complex with alendronate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-AMINO-1-HYDROXYBUTANE-1,1-DIYLDIPHOSPHONATE, CHLORIDE ION, ... | | Authors: | Dimos, N, Driller, R, Loll, B. | | Deposit date: | 2020-10-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The Impression of a Nonexisting Catalytic Effect: The Role of CotB2 in Guiding the Complex Biosynthesis of Cyclooctat-9-en-7-ol.
J.Am.Chem.Soc., 142, 2020
|
|
2LWW
 
 | | NMR structure of RelA-TAD/CBP-TAZ1 complex | | Descriptor: | CREB-binding protein, Nuclear transcription factor RelA, ZINC ION | | Authors: | Mukherjee, S.P, Ghosh, G, Wright, P.E. | | Deposit date: | 2012-08-07 | | Release date: | 2013-09-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Analysis of the RelA:CBP/p300 Interaction Reveals Its Involvement in NF-kappa B-Driven Transcription.
Plos Biol., 11, 2013
|
|
5L1P
 
 | | X-ray Structure of Cytochrome P450 PntM with Pentalenolactone | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase, pentalenolactone | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
2ZVU
 
 | | Crystal structure of rat heme oxygenase-1 in complex with ferrous verdoheme | | Descriptor: | 5-OXA-PROTOPORPHYRIN IX CONTAINING FE, FORMIC ACID, Heme oxygenase 1 | | Authors: | Sato, H, Sugishima, M, Fukuyama, K, Noguchi, M. | | Deposit date: | 2008-11-21 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of rat haem oxygenase-1 in complex with ferrous verdohaem: presence of a hydrogen-bond network on the distal side
Biochem.J., 419, 2009
|
|
7KB8
 
 | | Co-crystal structure of alpha glucosidase with compound 8 | | Descriptor: | (1S,2S,3R,4S,5S)-1-(hydroxymethyl)-5-[(4-{[2-nitro-4-(triazan-1-yl)phenyl]amino}butyl)amino]cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-10-01 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
2ZI8
 
 | | Crystal structure of the HsaC extradiol dioxygenase from M. tuberculosis in complex with 3,4-dihydroxy-9,10-seconandrost-1,3,5(10)-triene-9,17-dione (DHSA) | | Descriptor: | 3,4-dihydroxy-9,10-secoandrosta-1(10),2,4-triene-9,17-dione, FE (II) ION, PROBABLE BIPHENYL-2,3-DIOL 1,2-DIOXYGENASE BPHC | | Authors: | D'Angelo, I, Yam, K.C, Eltis, L.D, Strynadka, N. | | Deposit date: | 2008-02-13 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Studies of a ring-cleaving dioxygenase illuminate the role of cholesterol metabolism in the pathogenesis of Mycobacterium tuberculosis.
Plos Pathog., 5, 2009
|
|
7SXO
 
 | | Yeast Lon (PIM1) with endogenous substrate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Lon protease homolog, ... | | Authors: | Yang, J, Song, A.S, Wiseman, R.L, Lander, G.C. | | Deposit date: | 2021-11-24 | | Release date: | 2022-01-12 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of hexameric yeast Lon protease (PIM1) highlights the importance of conserved structural elements.
J.Biol.Chem., 298, 2022
|
|
7KAD
 
 | | Co-crystal structure of alpha glucosidase with compound 6 | | Descriptor: | (1S,2S,3R,4S,5S)-1-(hydroxymethyl)-5-[(6-{[2-nitro-4-(1H-1,2,3-triazol-1-yl)phenyl]amino}hexyl)amino]cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-09-30 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
3IFC
 
 | | Human muscle fructose-1,6-bisphosphatase E69Q mutant in complex with AMP and alpha fructose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase isozyme 2, ... | | Authors: | Kolodziejczyk, R, Zarzycki, M, Jaskolski, M, Dzugaj, A. | | Deposit date: | 2009-07-24 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of E69Q mutant of human muscle fructose-1,6-bisphosphatase
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2HHA
 
 | | The structure of DPP4 in complex with an oxadiazole inhibitor | | Descriptor: | (2S,3S)-3-{3-[4-(METHYLSULFONYL)PHENYL]-1,2,4-OXADIAZOL-5-YL}-1-OXO-1-PYRROLIDIN-1-YLBUTAN-2-AMINE, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2006-06-28 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of potent, selective, and orally bioavailable oxadiazole-based dipeptidyl peptidase IV inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2MAK
 
 | |
7PGC
 
 | |
