4LB7
 
 | |
4JW0
 
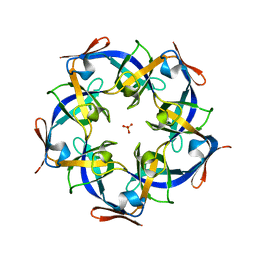 | | Structure of Gloeobacter violaceus CcmL | | Descriptor: | Carbon dioxide concentrating mechanism protein, SULFATE ION | | Authors: | Sutter, M, Kerfeld, C.A. | | Deposit date: | 2013-03-26 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Two new high-resolution crystal structures of carboxysome pentamer proteins reveal high structural conservation of CcmL orthologs among distantly related cyanobacterial species.
Photosynth.Res., 118, 2013
|
|
2J9P
 
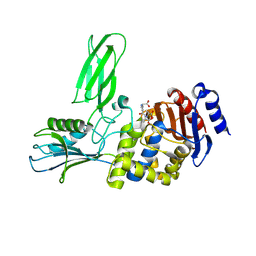 | | Crystal structure of the Bacillus subtilis PBP4a, and its complex with a peptidoglycan mimetic peptide. | | Descriptor: | (2R)-2-AMINO-7-{[(1R)-1-CARBOXYETHYL]AMINO}-7-OXOHEPTANOIC ACID, D-ALANINE, D-alanyl-D-alanine carboxypeptidase DacC | | Authors: | Sauvage, E, Herman, R, Kerff, F, Duez, C, Charlier, P. | | Deposit date: | 2006-11-15 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Bacillus subtilis penicillin-binding protein 4a, and its complex with a peptidoglycan mimetic peptide.
J. Mol. Biol., 371, 2007
|
|
4PRG
 
 | | 0072 PARTIAL AGONIST PPAR GAMMA COCRYSTAL | | Descriptor: | (+/-)(2S,5S)-3-(4-(4-CARBOXYPHENYL)BUTYL)-2-HEPTYL-4-OXO-5-THIAZOLIDINE, PROTEIN (PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR GAMMA) | | Authors: | Milburn, M.V. | | Deposit date: | 1999-05-07 | | Release date: | 1999-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A peroxisome proliferator-activated receptor gamma ligand inhibits adipocyte differentiation.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
4LG2
 
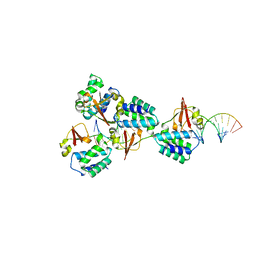 | | Crystal structure of Reston Ebola virus VP35 RNA binding domain bound to 12-bp dsRNA | | Descriptor: | Polymerase cofactor, dsRNA | | Authors: | Bale, S, Julien, J.-P, Bornholdt, Z.A, Krois, A.S, Wilson, I.A, Saphire, E.O. | | Deposit date: | 2013-06-27 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ebolavirus VP35 coats the backbone of double-stranded RNA for interferon antagonism.
J.Virol., 87, 2013
|
|
4LBF
 
 | |
5TOO
 
 | | Crystal structure of alkaline phosphatase PafA T79S, N100A, K162A, R164A mutant | | Descriptor: | Alkaline phosphatase PafA, CHLORIDE ION, ZINC ION | | Authors: | Lyubimov, A.Y, Sunden, F, AlSadhan, I, Herschlag, D. | | Deposit date: | 2016-10-18 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Differential catalytic promiscuity of the alkaline phosphatase superfamily bimetallo core reveals mechanistic features underlying enzyme evolution.
J. Biol. Chem., 292, 2017
|
|
2NTF
 
 | |
5TJ3
 
 | | Crystal structure of wild type alkaline phosphatase PafA to 1.7A resolution | | Descriptor: | Alkaline phosphatase PafA, ZINC ION | | Authors: | Lyubimov, A.Y, Sunden, F, Ressl, S, Herschlag, D. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanistic and Evolutionary Insights from Comparative Enzymology of Phosphomonoesterases and Phosphodiesterases across the Alkaline Phosphatase Superfamily.
J.Am.Chem.Soc., 138, 2016
|
|
2V7M
 
 | | PrnB 7-Cl-D-tryptophan complex | | Descriptor: | 7-CL-D-TRYPTOPHAN, PHOSPHATE ION, PRNB, ... | | Authors: | Naismith, J.H. | | Deposit date: | 2007-07-30 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Second Enzyme in Pyrrolnitrin Biosynthetic Pathway is Related to the Heme-Dependent Dioxygenase Superfamily
Biochemistry, 46, 2007
|
|
2V7K
 
 | | PrnB D-tryptophan complex | | Descriptor: | D-TRYPTOPHAN, PRNB, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Naismith, J.H. | | Deposit date: | 2007-07-30 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Second Enzyme in Pyrrolnitrin Biosynthetic Pathway is Related to the Heme-Dependent Dioxygenase Superfamily
Biochemistry, 46, 2007
|
|
2PNB
 
 | | STRUCTURE OF AN SH2 DOMAIN OF THE P85 ALPHA SUBUNIT OF PHOSPHATIDYLINOSITOL-3-OH KINASE | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT N-TERMINAL SH2 DOMAIN | | Authors: | Booker, G.W, Breeze, A.L, Downing, A.K, Panayotou, G, Gout, I, Waterfield, M.D, Campbell, I.D. | | Deposit date: | 1992-06-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of an SH2 domain of the p85 alpha subunit of phosphatidylinositol-3-OH kinase.
Nature, 358, 1992
|
|
2PYR
 
 | | PHOTOACTIVE YELLOW PROTEIN, 1 NANOSECOND INTERMEDIATE (287K) | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Perman, B, Srajer, V, Ren, Z, Teng, T.Y, Pradervand, C, Ursby, T, Bourgeois, D, Schotte, F, Wulff, M, Kort, R, Hellingwerf, K, Moffat, K. | | Deposit date: | 1998-03-04 | | Release date: | 1999-04-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Energy transduction on the nanosecond time scale: early structural events in a xanthopsin photocycle.
Science, 279, 1998
|
|
2WM8
 
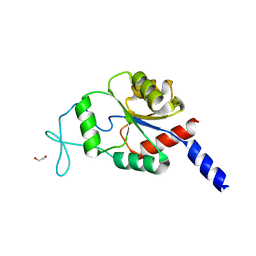 | | Crystal structure of human magnesium-dependent phosphatase 1 of the haloacid dehalogenase superfamily (MGC5987) | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM-DEPENDENT PHOSPHATASE 1 | | Authors: | Yue, W.W, Shafqat, N, Pike, A.C.W, Chaikuad, A, Bray, J.E, Pilka, E.W, Burgess-Brown, N, Hapka, E, Filippakopoulos, P, von Delft, F, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Human Magnesium-Dependent Phosphatase 1 of the Haloacid Dehalogenase Superfamily (Mgc5987)
To be Published
|
|
2KI5
 
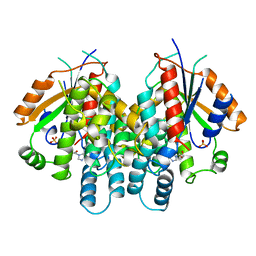 | | HERPES SIMPLEX TYPE-1 THYMIDINE KINASE IN COMPLEX WITH THE DRUG ACICLOVIR AT 1.9A RESOLUTION | | Descriptor: | 9-HYROXYETHOXYMETHYLGUANINE, PROTEIN (THYMIDINE KINASE), SULFATE ION | | Authors: | Bennett, M.S, Wien, F, Champness, J.N, Batuwangala, T, Rutherford, T, Summers, W.C, Sun, H, Wright, G, Sanderson, M.R. | | Deposit date: | 1999-02-12 | | Release date: | 1999-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure to 1.9 A resolution of a complex with herpes simplex virus type-1 thymidine kinase of a novel, non-substrate inhibitor: X-ray crystallographic comparison with binding of aciclovir.
FEBS Lett., 443, 1999
|
|
2PNI
 
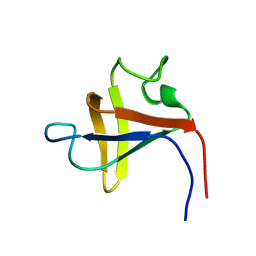 | | SOLUTION STRUCTURE AND LIGAND-BINDING SITE OF THE SH3 DOMAIN OF THE P85ALPHA SUBUNIT OF PHOSPHATIDYLINOSITOL 3-KINASE | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT SH3 DOMAIN | | Authors: | Booker, G.W, Gout, I, Downing, A.K, Driscoll, P.C, Boyd, J, Waterfield, M.D, Campbell, I.D. | | Deposit date: | 1993-07-19 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and ligand-binding site of the SH3 domain of the p85 alpha subunit of phosphatidylinositol 3-kinase.
Cell(Cambridge,Mass.), 73, 1993
|
|
2GEK
 
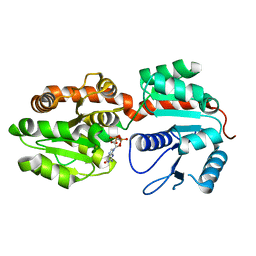 | | Crystal Structure of phosphatidylinositol mannosyltransferase (PimA) from Mycobacterium smegmatis in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, PHOSPHATIDYLINOSITOL MANNOSYLTRANSFERASE (PimA) | | Authors: | Guerin, M.E, Buschiazzo, A, Kordulakova, J, Jackson, M, Alzari, P.M. | | Deposit date: | 2006-03-20 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria.
J.Biol.Chem., 282, 2007
|
|
2PY8
 
 | | RbcX | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CHLORIDE ION, Hypothetical protein rbcX | | Authors: | Tanaka, S, Sawaya, M.R, Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 2007-05-15 | | Release date: | 2007-10-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the RuBisCO chaperone RbcX from Synechocystis sp. PCC6803.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2PNA
 
 | | STRUCTURE OF AN SH2 DOMAIN OF THE P85 ALPHA SUBUNIT OF PHOSPHATIDYLINOSITOL-3-OH KINASE | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT N-TERMINAL SH2 DOMAIN | | Authors: | Booker, G.W, Breeze, A.L, Downing, A.K, Panayotou, G, Gout, I, Waterfield, M.D, Campbell, I.D. | | Deposit date: | 1992-06-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of an SH2 domain of the p85 alpha subunit of phosphatidylinositol-3-OH kinase.
Nature, 358, 1992
|
|
4N1H
 
 | | Structure of a single-domain camelid antibody fragment cAb-F11N in complex with the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | Beta-lactamase, Camelid heavy-chain antibody variable fragment cAb-F11N | | Authors: | Pain, C, Kerff, F, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To be Published
|
|
2L87
 
 | | The 27-residue N-terminus CCR5-peptide in a ternary complex with HIV-1 gp120 and a CD4-mimic peptide | | Descriptor: | C-C chemokine receptor type 5 | | Authors: | Schnur, E, Noah, E, Ayzenshtat, I, Sargsyan, H, Inui, T, Ding, F.X, Arshava, B, Sagi, Y, Kessler, N, Levy, R, Scherf, T, Naider, F, Anglister, J. | | Deposit date: | 2011-01-06 | | Release date: | 2011-07-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Conformation and Orientation of a 27-Residue CCR5 Peptide in a Ternary Complex with HIV-1 gp120 and a CD4-Mimic Peptide.
J.Mol.Biol., 410, 2011
|
|
2QZ6
 
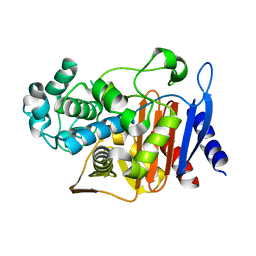 | | First crystal structure of a psychrophile class C beta-lactamase | | Descriptor: | Beta-lactamase | | Authors: | Michaux, C, Massant, J, Kerff, F, Charlier, P, Wouters, J. | | Deposit date: | 2007-08-16 | | Release date: | 2008-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of a cold-adapted class C beta-lactamase
Febs J., 275, 2008
|
|
2QW7
 
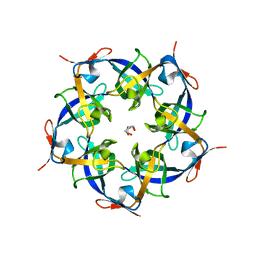 | | Carboxysome Subunit, CcmL | | Descriptor: | Carbon dioxide concentrating mechanism protein ccmL, GLYCEROL | | Authors: | Tanaka, S, Sawaya, M.R, Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 2007-08-09 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Atomic-level models of the bacterial carboxysome shell.
Science, 319, 2008
|
|
2BH7
 
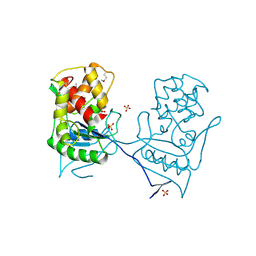 | | Crystal structure of a SeMet derivative of AmiD at 2.2 angstroms | | Descriptor: | N-ACETYLMURAMOYL-L-ALANINE AMIDASE, SULFATE ION, ZINC ION | | Authors: | Petrella, S, Herman, R, Sauvage, E, Genereux, C, Pennartz, A, Joris, B, Charlier, P. | | Deposit date: | 2005-01-07 | | Release date: | 2006-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Specific Structural Features of the N-Acetylmuramoyl-L-Alanine Amidase Amid from Escherichia Coli and Mechanistic Implications for Enzymes of This Family.
J.Mol.Biol., 397, 2010
|
|
2RQH
 
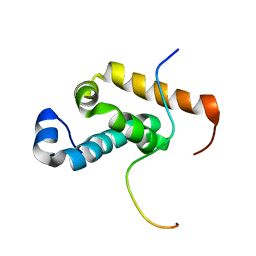 | | Structure of GSPT1/ERF3A-PABC | | Descriptor: | G1 to S phase transition 1, Polyadenylate-binding protein 1 | | Authors: | Osawa, M, Nakanishi, T, Hosoda, N, Uchida, S, Hoshino, T, Katada, I, Shimada, I. | | Deposit date: | 2009-05-08 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Eukaryotic Translation Termination Factor Gspt/Erf3 Recognizes Pabp with Chemical Exchange Using Two Overlapping Motifs
To be Published
|
|
