2OU0
 
 | |
7C2B
 
 | | Crystal structure of ferredoxin: thioredoxin reductase and thioredoxin f2 complex | | Descriptor: | Ferredoxin-thioredoxin reductase catalytic chain, chloroplastic, Ferredoxin-thioredoxin reductase variable chain, ... | | Authors: | Kurisu, G, Juniar, L, Tanaka, H. | | Deposit date: | 2020-05-07 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7949 Å) | | Cite: | Structural basis for thioredoxin isoform-based fine-tuning of ferredoxin-thioredoxin reductase activity.
Protein Sci., 29, 2020
|
|
3TRP
 
 | | Crystal structure of recombinant rabbit skeletal calsequestrin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sanchez, E.J, Lewis, K.M, Munske, G.R, Nissen, M.S, Kang, C. | | Deposit date: | 2011-09-09 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8817 Å) | | Cite: | High-capacity Ca2+ Binding of Human Skeletal Calsequestrin.
J.Biol.Chem., 287, 2012
|
|
6CVC
 
 | |
6CVM
 
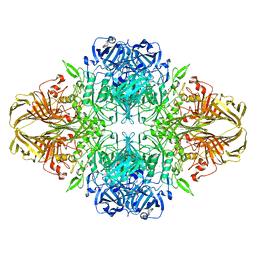 | | Atomic resolution cryo-EM structure of beta-galactosidase | | Descriptor: | 2-phenylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, MAGNESIUM ION, ... | | Authors: | Subramaniam, S, Bartesaghi, A, Banerjee, S, Zhu, X, Milne, J.L.S. | | Deposit date: | 2018-03-28 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Atomic Resolution Cryo-EM Structure of beta-Galactosidase.
Structure, 26, 2018
|
|
2JB3
 
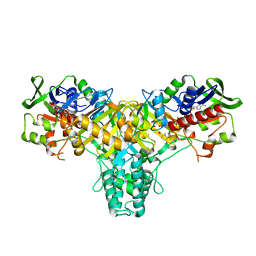 | | The structure of L-amino acid oxidase from Rhodococcus opacus in complex with o-aminobenzoate | | Descriptor: | 2-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, L-AMINO ACID OXIDASE | | Authors: | Faust, A, Niefind, K, hummel, W, Schomburg, D. | | Deposit date: | 2006-12-01 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of a Bacterial L-Amino Acid Oxidase from Rhodococcus Opacus Gives New Evidence for the Hydride Mechanism for Dehydrogenation
J.Mol.Biol., 367, 2007
|
|
7BYW
 
 | |
2OYL
 
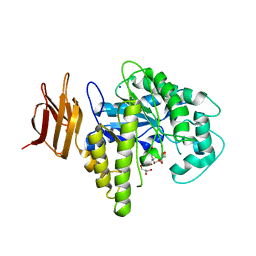 | | Endo-glycoceramidase II from Rhodococcus sp.: cellobiose-like imidazole complex | | Descriptor: | (5R,6R,7R,8S)-7,8-dihydroxy-5-(hydroxymethyl)-5,6,7,8-tetrahydroimidazo[1,2-a]pyridin-6-yl beta-D-glucopyranoside, Endoglycoceramidase II, GLYCEROL, ... | | Authors: | Caines, M.E.C, Strynadka, N.C.J. | | Deposit date: | 2007-02-22 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of glycosidase inhibition by five-membered iminocyclitols: the clan a glycoside hydrolase endoglycoceramidase as a model system.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
2XVQ
 
 | | Human serum albumin complexed with dansyl-L-sarcosine | | Descriptor: | DANSYL-L-SARCOSINE, SERUM ALBUMIN | | Authors: | Ryan, A.J, Curry, S. | | Deposit date: | 2010-10-27 | | Release date: | 2010-11-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis of Binding of Fluorescent, Site-Specific Dansylated Amino Acids to Human Serum Albumin.
J.Struct.Biol., 174, 2011
|
|
7BZC
 
 | | Crystal structure of plant sesterterpene synthase AtTPS18 complexed with farnesyl thiolodiphosphate (FSPP) | | Descriptor: | MAGNESIUM ION, S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE, Terpenoid synthase 18 | | Authors: | Li, J.X, Wang, G.D, Zhang, P. | | Deposit date: | 2020-04-27 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Molecular Basis for Sesterterpene Diversity Produced by Plant Terpene Synthases.
Plant Commun., 1, 2020
|
|
6CXV
 
 | | Structure of the S167H mutant of human indoleamine 2,3 dioxygenase in complex with tryptophan and cyanide | | Descriptor: | 2-(1H-indol-3-yl)ethanol, CYANIDE ION, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Lewis-Ballester, A, Yeh, S.-R, Karkashon, S, Batabyal, D, Poulos, T.L. | | Deposit date: | 2018-04-04 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Inhibition Mechanisms of Human Indoleamine 2,3 Dioxygenase 1.
J. Am. Chem. Soc., 140, 2018
|
|
2VDD
 
 | | Crystal Structure of the Open State of TolC Outer Membrane Component of Mutlidrug Efflux Pumps | | Descriptor: | CHLORIDE ION, OUTER MEMBRANE PROTEIN TOLC, POTASSIUM ION | | Authors: | Bavro, V.N, Pietras, Z, Furnham, N, Perez-Cano, L, Fernandez-Recio, J, Pei, X.Y, Truer, R, Misra, R, Luisi, B. | | Deposit date: | 2007-10-04 | | Release date: | 2008-04-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Assembly and Channel Opening in a Bacterial Drug Efflux Machine.
Mol.Cell, 30, 2008
|
|
4YXM
 
 | | Structure of Thermotoga maritima DisA D75N mutant with reaction product c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, (4S)-2-METHYL-2,4-PENTANEDIOL, DNA integrity scanning protein DisA | | Authors: | Mueller, M, Deimling, T, Hopfner, K.-P, Witte, G. | | Deposit date: | 2015-03-23 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural analysis of the diadenylate cyclase reaction of DNA-integrity scanning protein A (DisA) and its inhibition by 3'-dATP.
Biochem.J., 469, 2015
|
|
6SU8
 
 | | Highly thermostable endoglucanase Cel7B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucanase, ... | | Authors: | Schiano-di-Cola, C, Morth, J.P, Westh, P, Borch, K. | | Deposit date: | 2019-09-13 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural and biochemical characterization of a family 7 highly thermostable endoglucanase from the fungus Rasamsonia emersonii.
Febs J., 287, 2020
|
|
2OXB
 
 | | Crystal structure of a cell-wall invertase (E203Q) from Arabidopsis thaliana in complex with sucrose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-fructofuranosidase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Lammens, W, Le Roy, K, Van Laere, A, Van den Ende, W, Rabijns, A. | | Deposit date: | 2007-02-20 | | Release date: | 2008-01-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An alternate sucrose binding mode in the E203Q Arabidopsis invertase mutant: An X-ray crystallography and docking study.
Proteins, 71, 2007
|
|
2ONP
 
 | | Arg475Gln Mutant of Human Mitochondrial Aldehyde Dehydrogenase, complexed with NAD+ | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde dehydrogenase, GUANIDINE, ... | | Authors: | Larson, H.N, Hurley, T.D. | | Deposit date: | 2007-01-24 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional consequences of coenzyme binding to the inactive asian variant of mitochondrial aldehyde dehydrogenase: roles of residues 475 and 487.
J.Biol.Chem., 282, 2007
|
|
2I30
 
 | | Human serum albumin complexed with myristate and salicylic acid | | Descriptor: | 2-HYDROXYBENZOIC ACID, MYRISTIC ACID, Serum albumin | | Authors: | Yang, F, Bian, C, Zhu, L, Zhao, G, Huang, Z, Huang, M. | | Deposit date: | 2006-08-17 | | Release date: | 2006-12-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Effect of human serum albumin on drug metabolism: Structural evidence of esterase activity of human serum albumin
J.Struct.Biol., 157, 2007
|
|
4YZT
 
 | | Crystal structure of a tri-modular GH5 (subfamily 4) endo-beta-1, 4-glucanase from Bacillus licheniformis complexed with cellotetraose | | Descriptor: | 1,2-ETHANEDIOL, Cellulose hydrolase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Liberato, M.V, Popov, A, Polikarpov, I. | | Deposit date: | 2015-03-25 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Molecular characterization of a family 5 glycoside hydrolase suggests an induced-fit enzymatic mechanism.
Sci Rep, 6, 2016
|
|
2HYU
 
 | | Human Annexin A2 with heparin tetrasaccharide bound | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Annexin A2, CALCIUM ION | | Authors: | Shao, C, Head, J.F, Seaton, B.A. | | Deposit date: | 2006-08-07 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystallographic Analysis of Calcium-dependent Heparin Binding to Annexin A2.
J.Biol.Chem., 281, 2006
|
|
4Z2U
 
 | |
4AXK
 
 | | CRYSTAL STRUCTURE OF subHisA from the thermophile Corynebacterium efficiens | | Descriptor: | 1-(5-PHOSPHORIBOSYL)-5-((5'-PHOSPHORIBOSYLAMINO) METHYLIDENEAMINO)IMIDAZOLE-4-CARBOXAMIDE ISOMERASE, GLYCEROL | | Authors: | Noda-Garcia, L, Camacho-Zarco, A.R, Medina-Ruiz, S, Verduzco-Castro, E.A, Gaytan, P, Carrillo-Tripp, M, Fulop, V, Barona-Gomez, F. | | Deposit date: | 2012-06-13 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Evolution of Substrate Specificity in a Recipient'S Enzyme Following Horizontal Gene Transfer.
Mol.Biol.Evol., 30, 2013
|
|
6CXT
 
 | | Crystal structure of FAD-dependent dehydrogenase | | Descriptor: | 1,2-ETHANEDIOL, Alpha/beta hydrolase fold protein, Butyryl-CoA dehydrogenase, ... | | Authors: | Agarwal, V. | | Deposit date: | 2018-04-04 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into Thiotemplated Pyrrole Biosynthesis Gained from the Crystal Structure of Flavin-Dependent Oxidase in Complex with Carrier Protein.
Biochemistry, 58, 2019
|
|
2OQ2
 
 | | Crystal structure of yeast PAPS reductase with PAP, a product complex | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Phosphoadenosine phosphosulfate reductase | | Authors: | Yu, Z, Fisher, A.J. | | Deposit date: | 2007-01-31 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae 3'-phosphoadenosine-5'-phosphosulfate reductase complexed with adenosine 3',5'-bisphosphate.
Biochemistry, 47, 2008
|
|
6CY9
 
 | | SA11 Rotavirus NSP2 with disulfide bridge | | Descriptor: | MAGNESIUM ION, Non-structural protein 2 | | Authors: | Anish, R, Hu, L, Sankaran, B, Prasad, B.V.V. | | Deposit date: | 2018-04-05 | | Release date: | 2018-12-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.615 Å) | | Cite: | Phosphorylation cascade regulates the formation and maturation of rotaviral replication factories.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CWJ
 
 | | Crystal structures of cyanuric acid hydrolase from Moorella thermoacetica complexed with 1,3-Acetone Dicarboxylic Acid | | Descriptor: | 1,3-PROPANDIOL, 3-oxopentanedioic acid, ACETATE ION, ... | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2018-03-30 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Crystal structures of Moorella thermoacetica cyanuric acid hydrolase reveal conformational flexibility and asymmetry important for catalysis.
Plos One, 14, 2019
|
|
