6ETD
 
 | |
4DH8
 
 | | Room temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, AMP-PNP and IP20 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DH7
 
 | | Low temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, AMP-PNP and IP20' | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6AIB
 
 | | Crystal structures of the N-terminal RecA-like domain 1 of Staphylococcus aureus DEAD-box Cold shock RNA helicase CshA | | Descriptor: | DEAD-box ATP-dependent RNA helicase CshA | | Authors: | Chengliang, W, Tian, T, Xiaobao, C, Xuan, Z, Jianye, Z. | | Deposit date: | 2018-08-22 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of the N-terminal domain of the Staphylococcus aureus DEAD-box RNA helicase CshA and its complex with AMP
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
3A7A
 
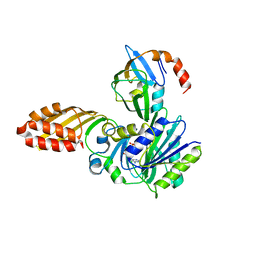 | | Crystal structure of E. coli lipoate-protein ligase A in complex with octyl-amp and apoH-protein | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glycine cleavage system H protein, Lipoate-protein ligase A, ... | | Authors: | Fujiwara, K, Hosaka, H, Nakagawa, A. | | Deposit date: | 2009-09-20 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Global conformational change associated with the two-step reaction catalyzed by Escherichia coli lipoate-protein ligase A.
J.Biol.Chem., 285, 2010
|
|
3SZG
 
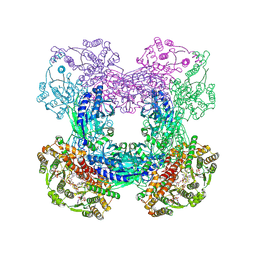 | | Crystal structure of C176A glutamine-dependent NAD+ synthetase from M. tuberculosis bound to AMP/PPi and NaAD+ | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, Glutamine-dependent NAD(+) synthetase, ... | | Authors: | Chuenchor, W, Doukov, T, Gerratana, B. | | Deposit date: | 2011-07-19 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Regulation of the intersubunit ammonia tunnel in Mycobacterium tuberculosis glutamine-dependent NAD+ synthetase.
Biochem.J., 443, 2012
|
|
3SYT
 
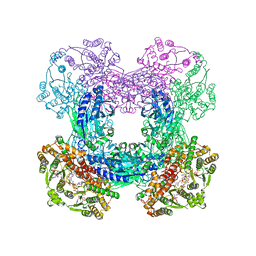 | | Crystal structure of glutamine-dependent NAD+ synthetase from M. tuberculosis bound to AMP/PPi, NAD+, and glutamate | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Chuenchor, W, Gerratana, B. | | Deposit date: | 2011-07-18 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6511 Å) | | Cite: | Regulation of the intersubunit ammonia tunnel in Mycobacterium tuberculosis glutamine-dependent NAD+ synthetase.
Biochem.J., 443, 2012
|
|
3A7R
 
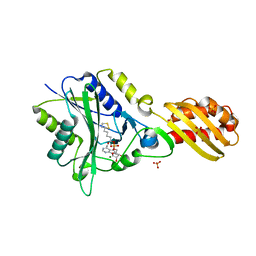 | |
5GZW
 
 | | Crystal structure of AmpC BER adenylylated by acetyl-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Beta-lactamase, SULFATE ION | | Authors: | An, Y.J, Cha, S.S. | | Deposit date: | 2016-10-02 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Structural and mechanistic insights into the inhibition of class C beta-lactamases through the adenylylation of the nucleophilic serine.
J.Antimicrob.Chemother., 72, 2017
|
|
1J1Z
 
 | |
5YFU
 
 | | Crystal structure of ribose-1,5-bisphosphate isomerase from Pyrococcus horikoshii OT3 in complex with ribulose-1,5-bisphosphate and AMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE MONOPHOSPHATE, POTASSIUM ION, ... | | Authors: | Gogoi, P, Kanaujia, S.P. | | Deposit date: | 2017-09-22 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A presumed homologue of the regulatory subunits of eIF2B functions as ribose-1,5-bisphosphate isomerase in Pyrococcus horikoshii OT3.
Sci Rep, 8, 2018
|
|
5YG8
 
 | | Crystal structure of ribose-1,5-bisphosphate isomerase from Pyrococcus horikoshii OT3 in complex with ribulose-1,5-bisphosphate, AMP and GMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE MONOPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Gogoi, P, Kanaujia, S.P. | | Deposit date: | 2017-09-22 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A presumed homologue of the regulatory subunits of eIF2B functions as ribose-1,5-bisphosphate isomerase in Pyrococcus horikoshii OT3.
Sci Rep, 8, 2018
|
|
3TY9
 
 | | Crystal Structure of C. Thermocellum PNKP Ligase Domain AMP-Adenylate | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Smith, P, Wang, L, Shuman, S. | | Deposit date: | 2011-09-24 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | The adenylyltransferase domain of bacterial Pnkp defines a unique RNA ligase family.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5BPH
 
 | | Crystal structure of AMP complexed D-alanine-D-alanine ligase(DDL) from Yersinia pestis | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, D-alanine--D-alanine ligase, ... | | Authors: | Tran, H.T, Kang, L.W, Hong, M.K. | | Deposit date: | 2015-05-28 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of D-alanine-D-alanine ligase from Yersinia pestis: nucleotide phosphate recognition by the serine loop.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4R9U
 
 | | Structure of vitamin B12 transporter BtuCD in a nucleotide-bound outward facing state | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Korkhov, V.M, Mireku, S.A, Veprintsev, D.B, Locher, K.P. | | Deposit date: | 2014-09-08 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.785 Å) | | Cite: | Structure of AMP-PNP-bound BtuCD and mechanism of ATP-powered vitamin B12 transport by BtuCD-F.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4S32
 
 | |
6A06
 
 | | Structure of pSTING complex | | Descriptor: | SULFATE ION, Stimulator of interferon genes protein, cGAMP | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
2YPE
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation H309S, crystallized with 2',3'- cyclic AMP | | Descriptor: | 2', 3'-CYCLIC-NUCLEOTIDE 3'-PHOSPHODIESTERASE, 2',3'- cyclic AMP, ... | | Authors: | Myllykoski, M, Raasakka, A, Lehtimaki, M, Han, H, Kursula, P. | | Deposit date: | 2012-10-30 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic Analysis of the Reaction Cycle of 2',3'-Cyclic Nucleotide 3'-Phosphodiesterase, a Unique Member of the 2H Phosphoesterase Family
J.Mol.Biol., 425, 2013
|
|
1YXU
 
 | | Crystal Structure of Kinase Pim1 in Complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Kumar, A, Mandiyan, V, Suzuki, Y, Zhang, C, Rice, J, Tsai, J, Artis, D.R, Ibrahim, P, Bremer, R. | | Deposit date: | 2005-02-22 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structures of Proto-oncogene Kinase Pim1: A Target of Aberrant Somatic Hypermutations in Diffuse Large Cell Lymphoma.
J.Mol.Biol., 348, 2005
|
|
6ETF
 
 | |
6ETH
 
 | |
6GB0
 
 | |
6GBC
 
 | |
6GB9
 
 | |
5ZBK
 
 | |
