5OLR
 
 | | Rhamnogalacturonan lyase | | Descriptor: | CALCIUM ION, PHOSPHATE ION, Rhamnogalacturonan lyase, ... | | Authors: | Basle, A, Luis, A.S, Gilbert, H.J. | | Deposit date: | 2017-07-28 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides.
Nat Microbiol, 3, 2018
|
|
5O0A
 
 | | Crystal structure of Phosphopantetheine adenylyltransferase from Mycobacterium abcessus in complex with 5-methyl-1-phenyl-pyrazole-4-carboxylic acid (Fragment 1) | | Descriptor: | 5-methyl-1-phenyl-pyrazole-4-carboxylic acid, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, Kim, S.Y, Mendes, V, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2017-05-16 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Structural Biology and the Design of New Therapeutics: From HIV and Cancer to Mycobacterial Infections: A Paper Dedicated to John Kendrew.
J. Mol. Biol., 429, 2017
|
|
5O0F
 
 | | Crystal structure of Phosphopantetheine adenylyltransferase from Mycobacterium abcessus in complex with 3-(indol-3-yl)propanoic acid (Fragment 5) | | Descriptor: | INDOLYLPROPIONIC ACID, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, Kim, S.Y, Mendes, V, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2017-05-16 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Structural Biology and the Design of New Therapeutics: From HIV and Cancer to Mycobacterial Infections: A Paper Dedicated to John Kendrew.
J. Mol. Biol., 429, 2017
|
|
5O0N
 
 | | Deglycosylated Nogo Receptor with native disulfide structure 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Reticulon-4 receptor, ... | | Authors: | Pronker, M.F, Tas, R.P, Vlieg, H.C, Janssen, B.J.C. | | Deposit date: | 2017-05-16 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nogo Receptor crystal structures with a native disulfide pattern suggest a novel mode of self-interaction.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5OKJ
 
 | | Non-conservatively refined structure of Gan1D-WT, a putative 6-phospho-beta-galactosidase from Geobacillus stearothermophilus, in the C2 spacegroup | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, IMIDAZOLE, ... | | Authors: | Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2017-07-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural basis for enzyme bifunctionality - the case of Gan1D from Geobacillus stearothermophilus.
FEBS J., 284, 2017
|
|
5O0T
 
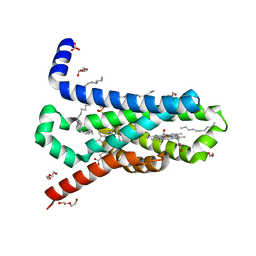 | | CRYSTAL STRUCTURE OF TRANS-MEMBRANE DOMAIN OF Cylindrospermum stagnale NADPH-OXIDASE 5 (NOX5) | | Descriptor: | DECANE, DI(HYDROXYETHYL)ETHER, EICOSANE, ... | | Authors: | Magnani, F, Nenci, S, Mattevi, A. | | Deposit date: | 2017-05-17 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures and atomic model of NADPH oxidase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5OLY
 
 | |
5O1P
 
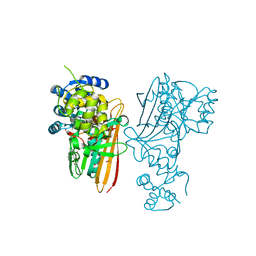 | | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase. | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde synthase, mitochondrial, ... | | Authors: | Kopec, J, Rembeza, E, Pena, I.A, Williams, E, Velupillai, S, Kupinska, K, Strain-Damerell, C, Goubin, S, Talon, R, Collins, P, Krojer, T, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, von Delft, F, Arruda, P, Yue, W.W. | | Deposit date: | 2017-05-18 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase.
To Be Published
|
|
5O1Y
 
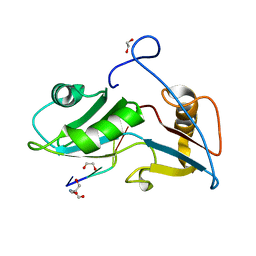 | | Structure of Nrd1 RNA binding domain in complex with RNA (GUAA) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Protein NRD1, ... | | Authors: | Franco-Echevarria, E, Perez-Canadillas, J.M, Gonzalez, B. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The structure of transcription termination factor Nrd1 reveals an original mode for GUAA recognition.
Nucleic Acids Res., 45, 2017
|
|
5O32
 
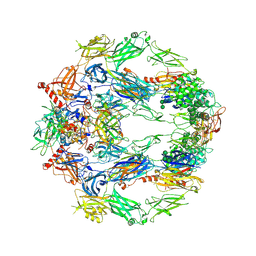 | | The structure of complement complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xue, X, Wu, J, Forneris, F, Gros, P. | | Deposit date: | 2017-05-23 | | Release date: | 2017-06-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4.206209 Å) | | Cite: | Regulator-dependent mechanisms of C3b processing by factor I allow differentiation of immune responses.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5O3W
 
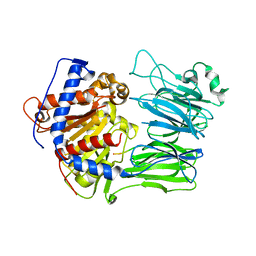 | | Structural characterization of the fast and promiscuous macrocyclase from plant - PCY1-S562A bound to Presegetalin A1 | | Descriptor: | MAGNESIUM ION, Peptide cyclase 1, Presegetalin A1, ... | | Authors: | Ludewig, H, Czekster, C.M, Bent, A.F, Naismith, J.H. | | Deposit date: | 2017-05-25 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the Fast and Promiscuous Macrocyclase from Plant PCY1 Enables the Use of Simple Substrates.
ACS Chem. Biol., 13, 2018
|
|
5O4T
 
 | |
5O7W
 
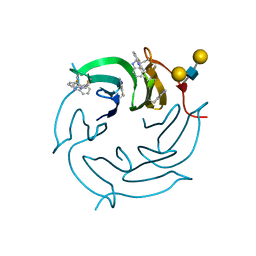 | | Crystal structure of the 4-FLUORO RSL lectin in complex with Lewis x tetrasaccharide | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Putative fucose-binding lectin protein, ... | | Authors: | Varrot, A. | | Deposit date: | 2017-06-09 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Effect of Noncanonical Amino Acids on Protein-Carbohydrate Interactions: Structure, Dynamics, and Carbohydrate Affinity of a Lectin Engineered with Fluorinated Tryptophan Analogs.
ACS Chem. Biol., 13, 2018
|
|
5O84
 
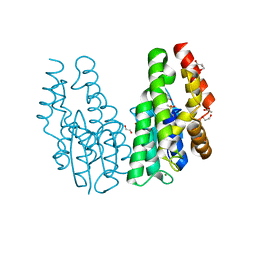 | | Glutathione S-transferase Tau 23 (partially oxidized) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FORMIC ACID, ... | | Authors: | Young, D.R, Van Molle, I, Tossounian, M, Messens, J. | | Deposit date: | 2017-06-12 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Disulfide bond formation protects Arabidopsis thaliana glutathione transferase tau 23 from oxidative damage.
Biochim. Biophys. Acta, 1862, 2018
|
|
5O8Y
 
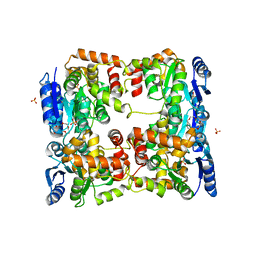 | | Conformational dynamism for DNA interaction in Salmonella typhimurium RcsB response regulator. | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, SULFATE ION, Transcriptional regulatory protein RcsB | | Authors: | Casino, P, Marina, A, Miguel-Romero, L, Huesa, J. | | Deposit date: | 2017-06-14 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational dynamism for DNA interaction in the Salmonella RcsB response regulator.
Nucleic Acids Res., 46, 2018
|
|
5O6E
 
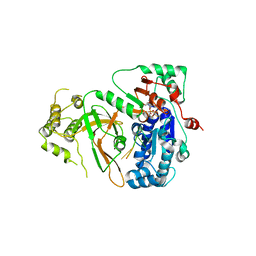 | | Structure of ScPif1 in complex with TTTGGGTT and ADP-AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase PIF1, CHLORIDE ION, ... | | Authors: | Lu, K.Y, Chen, W.F, Rety, S, Liu, N.N, Xu, X.G. | | Deposit date: | 2017-06-06 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.345 Å) | | Cite: | Insights into the structural and mechanistic basis of multifunctional S. cerevisiae Pif1p helicase.
Nucleic Acids Res., 46, 2018
|
|
5O9H
 
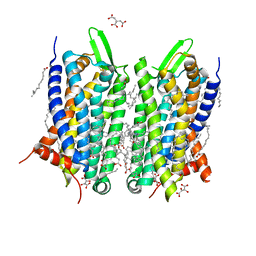 | | Crystal structure of thermostabilised human C5a anaphylatoxin chemotactic receptor 1 (C5aR) in complex with NDT9513727 | | Descriptor: | 1-(1,3-benzodioxol-5-yl)-~{N}-(1,3-benzodioxol-5-ylmethyl)-~{N}-[(3-butyl-2,5-diphenyl-imidazol-4-yl)methyl]methanamine, C5a anaphylatoxin chemotactic receptor 1, CITRIC ACID, ... | | Authors: | Robertson, N, Rappas, M, Dore, A.S, Brown, J, Bottegoni, G, Koglin, M, Cansfield, J, Jazayeri, A, Cooke, R.M, Marshall, F.H. | | Deposit date: | 2017-06-19 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the complement C5a receptor bound to the extra-helical antagonist NDT9513727.
Nature, 553, 2018
|
|
5O6S
 
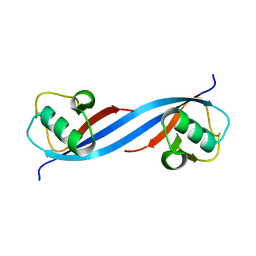 | |
5O75
 
 | | Ube4B U-box domain | | Descriptor: | SULFATE ION, Ubiquitin conjugation factor E4 B | | Authors: | Gabrielsen, M, Buetow, L, Nakasone, M.A, Huang, D.T. | | Deposit date: | 2017-06-08 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.483 Å) | | Cite: | A General Strategy for Discovery of Inhibitors and Activators of RING and U-box E3 Ligases with Ubiquitin Variants.
Mol. Cell, 68, 2017
|
|
5O79
 
 | | Klebsiella pneumoniae OmpK36 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, MAGNESIUM ION, OmpK36 | | Authors: | van den berg, B, Pathania, M. | | Deposit date: | 2017-06-08 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Getting Drugs into Gram-Negative Bacteria: Rational Rules for Permeation through General Porins.
Acs Infect Dis., 4, 2018
|
|
5O7U
 
 | |
5OCF
 
 | | Crystal structure of nitric oxide bound to three-domain heme-Cu nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, HEME C, NITRIC OXIDE, ... | | Authors: | Dong, J, Sasaki, D, Eady, R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Activation of redox tyrosine switch is required for ligand binding at the catalytic site in heme-cu nitrite reductases
To be published
|
|
5OCP
 
 | |
5OG0
 
 | | Crystal structure of human Alanine:Glyoxylate Aminotransferase major allele (AGT-Ma) at 2.5 Angstrom; internal aldimine with PLP in the active site | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Giardina, G, Cutruzzola, F, Borri Voltattorni, C, Cellini, B, Montioli, R. | | Deposit date: | 2017-07-11 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Radiation damage at the active site of human alanine:glyoxylate aminotransferase reveals that the cofactor position is finely tuned during catalysis.
Sci Rep, 7, 2017
|
|
5O9D
 
 | | Crystal structure of R. ruber ADH-A, mutant Y294F, W295A, Y54F, F43H, H39Y | | Descriptor: | (2~{S})-2-methylpentanedioic acid, Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Maurer, D, Hamnevik, E, Enugala, T.R, Widersten, M. | | Deposit date: | 2017-06-19 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Relaxation of nonproductive binding and increased rate of coenzyme release in an alcohol dehydrogenase increases turnover with a nonpreferred alcohol enantiomer.
FEBS J., 284, 2017
|
|
