1QUR
 
 | |
7NE4
 
 | | E125A mutant of oligopeptidase B from S. proteomaculans with modified hinge region | | Descriptor: | Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Nikolaeva, A.Y, Lazarenko, V.A, Dorovatovskiy, P.V, Vlaskina, A.V, Mikhailova, A.G, Rakitina, T.V, Timofeev, V.I. | | Deposit date: | 2021-02-03 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.717 Å) | | Cite: | First Crystal Structure of Bacterial Oligopeptidase B in an Intermediate State: The Roles of the Hinge Region Modification and Spermine.
Biology (Basel), 10, 2021
|
|
7NE5
 
 | | catalytically non active S532A mutant of oligopeptidase B from S. proteomaculans with modified hinge region | | Descriptor: | Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Nikolaeva, A.Y, Lazarenko, V.A, Dorovatovskiy, P.V, Vlaskina, A.V, Mikhailova, A.G, Rakitina, T.V, Timofeev, V.I. | | Deposit date: | 2021-02-03 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | First Crystal Structure of Bacterial Oligopeptidase B in an Intermediate State: The Roles of the Hinge Region Modification and Spermine.
Biology (Basel), 10, 2021
|
|
7OB1
 
 | | OLIGOPEPTIDASE B FROM S. PROTEOMACULANS WITH MODIFIED HINGE | | Descriptor: | Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Nikolaeva, A.Y, Lazarenko, V.A, Dorovatovskiy, P.V, Vlaskina, A.V, Korzhenevskiy, D.A, Mikhailova, A.G, Rakitina, T.V, Timofeev, V.I. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | First Crystal Structure of Bacterial Oligopeptidase B in an Intermediate State: The Roles of the Hinge Region Modification and Spermine.
Biology (Basel), 10, 2021
|
|
7SGJ
 
 | |
7SGI
 
 | |
7SGG
 
 | |
7SGK
 
 | |
7S3V
 
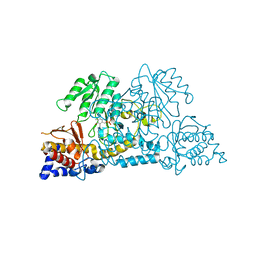 | |
1IAU
 
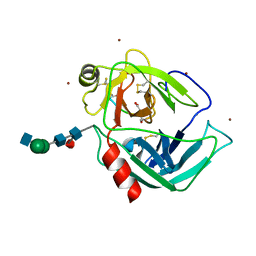 | | HUMAN GRANZYME B IN COMPLEX WITH AC-IEPD-CHO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GRANZYME B, ... | | Authors: | Rotonda, J, Garcia-Calvo, M, Bull, H.G, Geissler, W.M, McKeever, B.M, Willoughby, C.A, Thornberry, N.A, Becker, J.W. | | Deposit date: | 2001-03-23 | | Release date: | 2001-05-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The three-dimensional structure of human granzyme B compared to caspase-3, key mediators of cell death with cleavage specificity for aspartic acid in P1.
Chem.Biol., 8, 2001
|
|
4JSQ
 
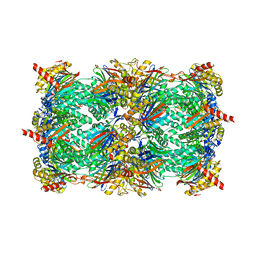 | | Yeast 20S proteasome in complex with the dimerized linear mimetic of TMC-95A - yCP:4e | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, ... | | Authors: | Desvergne, A, Genin, E, Marechal, X, Gallastegui, N, Dufau, L, Richy, N, Groll, M, Vidal, J, Reboud-Ravaux, M. | | Deposit date: | 2013-03-22 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dimerized linear mimics of a natural cyclopeptide (TMC-95A) are potent noncovalent inhibitors of the eukaryotic 20S proteasome
J.Med.Chem., 56, 2013
|
|
4JT0
 
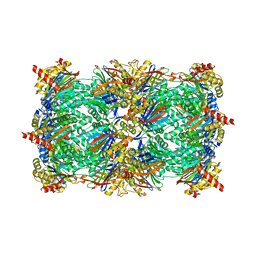 | | Yeast 20S proteasome in complex with the dimerized linear mimetic of TMC-95A - yCP:4a | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, ... | | Authors: | Desvergne, A, Genin, E, Marechal, X, Gallastegui, N, Dufau, L, Richy, N, Groll, M, Vidal, J, Reboud-Ravaux, M. | | Deposit date: | 2013-03-22 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Dimerized linear mimics of a natural cyclopeptide (TMC-95A) are potent noncovalent inhibitors of the eukaryotic 20S proteasome.
J.Med.Chem., 56, 2013
|
|
1HIA
 
 | | KALLIKREIN COMPLEXED WITH HIRUSTASIN | | Descriptor: | HIRUSTASIN, KALLIKREIN | | Authors: | Mittl, P, Di Marco, S, Gruetter, M. | | Deposit date: | 1996-12-12 | | Release date: | 1997-12-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A new structural class of serine protease inhibitors revealed by the structure of the hirustasin-kallikrein complex.
Structure, 5, 1997
|
|
4JSU
 
 | | Yeast 20S proteasome in complex with the dimerized linear mimetic of TMC-95A - yCP:3a | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, ... | | Authors: | Desvergne, A, Genin, E, Marechal, X, Gallastegui, N, Dufau, L, Richy, N, Groll, M, Vidal, J, Reboud-Ravaux, M. | | Deposit date: | 2013-03-22 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dimerized linear mimics of a natural cyclopeptide (TMC-95A) are potent noncovalent inhibitors of the eukaryotic 20S proteasome
J.Med.Chem., 56, 2013
|
|
1QIX
 
 | | Porcine pancreatic elastase complexed with human beta-casomorphin-7 | | Descriptor: | BETA-CASOMORPHIN-7, CALCIUM ION, ELASTASE, ... | | Authors: | Wilmouth, R.C, Clifton, I.J, Hajdu, J, Schofield, C.J. | | Deposit date: | 1999-06-18 | | Release date: | 1999-12-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Specific Acyl-Enzyme Complex Formed between Beta-Casomorphin-7 and Porcine Pancreatic Elastase
Nat.Struct.Biol., 4, 1997
|
|
6XHZ
 
 | | Alpha-lytic protease homolog N4 | | Descriptor: | N4: hypothetical protein, SULFATE ION | | Authors: | Nixon, C.F, Marqusee, S.M, Gee, C.L. | | Deposit date: | 2020-06-19 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Exploring the Evolutionary History of Kinetic Stability in the alpha-Lytic Protease Family.
Biochemistry, 60, 2021
|
|
6Y3B
 
 | | Crystal Structure of Unlinked NS2B-NS3 Protease from Zika Virus in Complex with Inhibitor MI-2110 | | Descriptor: | 1-[(8~{R},15~{S},18~{S})-15,18-bis(4-azanylbutyl)-4,7,14,17,20-pentakis(oxidanylidene)-3,6,13,16,19-pentazabicyclo[20.3.1]hexacosa-1(25),22(26),23-trien-8-yl]guanidine, GLYCEROL, Genome polyprotein, ... | | Authors: | Huber, S, Heine, A, Steinmetzer, T. | | Deposit date: | 2020-02-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-Based Macrocyclization of Substrate Analogue NS2B-NS3 Protease Inhibitors of Zika, West Nile and Dengue viruses.
Chemmedchem, 15, 2020
|
|
4Z8N
 
 | |
8OYH
 
 | | X-ray structure of furin (PCSK3) in complex with Guanidinomethyl-Phac-Can-Tle-Can-6-(aminomethyl)-3-amino-isoindol | | Descriptor: | CALCIUM ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Dahms, S.O, Brandstetter, H. | | Deposit date: | 2023-05-04 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-Based Design, Synthesis, and Characterization of Aminoisoindole-Derived Furin Inhibitors.
Chemmedchem, 19, 2024
|
|
2XW9
 
 | | Crystal Structure of Complement Factor D mutant S183A | | Descriptor: | COMPLEMENT FACTOR D, GLYCEROL | | Authors: | Forneris, F, Ricklin, D, Wu, J, Tzekou, A, Wallace, R.S, Lambris, J.D, Gros, P. | | Deposit date: | 2010-11-01 | | Release date: | 2011-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structures of C3B in Complex with Factors B and D Give Insight Into Complement Convertase Formation.
Science, 330, 2010
|
|
5A53
 
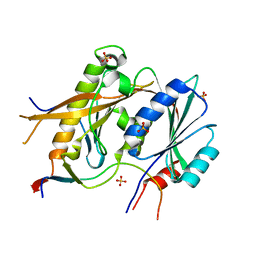 | | Crystal Structure of the Rpf2-Rrs1 complex | | Descriptor: | REGULATOR OF RIBOSOME BIOSYNTHESIS, RIBOSOME BIOGENESIS PROTEIN RPF2, SULFATE ION | | Authors: | Madru, C, Lebaron, S, Blaud, M, Delbos, L, Rety, S, Leulliot, N. | | Deposit date: | 2015-06-16 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Chaperoning 5S RNA Assembly.
Genes Dev., 29, 2015
|
|
7Q8I
 
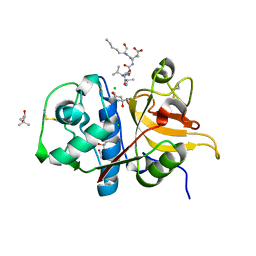 | | Peptide AVAEKQ in complex with human cathepsin V C25S mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, AVAEKQ peptide, CHLORIDE ION, ... | | Authors: | Loboda, J, Sosnowski, P, Tusar, L, Vidmar, R, Vizovisek, M, Horvat, J, Kosec, G, Impens, F, Demol, H, Turk, B, Gevaert, K, Turk, D. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Proteomic data and structure analysis combined reveal interplay of structural rigidity and flexibility on selectivity of cysteine cathepsins.
Commun Biol, 6, 2023
|
|
7Q8O
 
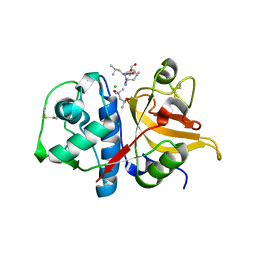 | | Peptide LLSGKE in complex with human cathepsin V C25S mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Cathepsin L2, ... | | Authors: | Loboda, J, Sosnowski, P, Tusar, L, Vidmar, R, Vizovisek, M, Horvat, J, Kosec, G, Impens, F, Demol, H, Turk, B, Gevaert, K, Turk, D. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Proteomic data and structure analysis combined reveal interplay of structural rigidity and flexibility on selectivity of cysteine cathepsins.
Commun Biol, 6, 2023
|
|
7Q8H
 
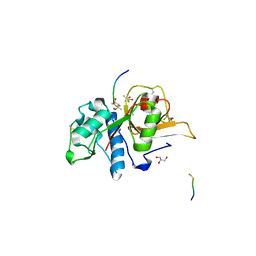 | | Peptide EVCKKKK in complex with human cathepsin V C25A mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Cathepsin L2, ... | | Authors: | Loboda, J, Sosnowski, P, Tusar, L, Vidmar, R, Vizovisek, M, Horvat, J, Kosec, G, Impens, F, Demol, H, Turk, B, Gevaert, K, Turk, D. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Proteomic data and structure analysis combined reveal interplay of structural rigidity and flexibility on selectivity of cysteine cathepsins.
Commun Biol, 6, 2023
|
|
7Q9H
 
 | | Peptide LLKAVAEKQ in complex with human cathepsin V C25A mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Cathepsin L2, ... | | Authors: | Loboda, J, Sosnowski, P, Tusar, L, Vidmar, R, Vizovisek, M, Horvat, J, Kosec, G, Impens, F, Demol, H, Turk, B, Gevaert, K, Turk, D. | | Deposit date: | 2021-11-12 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Proteomic data and structure analysis combined reveal interplay of structural rigidity and flexibility on selectivity of cysteine cathepsins.
Commun Biol, 6, 2023
|
|
