3HP5
 
 | | Crystal Structure of Human p38alpha complexed with a pyrimidopyridazinone compound | | Descriptor: | 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, 5-(2,6-dichlorophenyl)-2-[(2,4-difluorophenyl)sulfanyl]-6H-pyrimido[1,6-b]pyridazin-6-one, Mitogen-activated protein kinase 14 | | Authors: | Shieh, H.-S, Williams, J.M, Stegeman, R.A, Kurumbail, R.G. | | Deposit date: | 2009-06-03 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of N-substituted pyridinones as potent and selective inhibitors of p38 kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
7B4G
 
 | | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ). Human p53DBD-R282W mutant bound to DNA: R282W-MQ (II) | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Degtjarik, O, Rozenberg, H, Shakked, Z. | | Deposit date: | 2020-12-02 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ).
Nat Commun, 12, 2021
|
|
8KD8
 
 | |
5UIK
 
 | | X-ray structure of the FdtF formyltransferase from salmonella enteric O60 in complex with TDP-Fuc3N and folinic acid | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, Formyltransferase, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, ... | | Authors: | Woodford, C.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular architecture of an N-formyltransferase from Salmonella enterica O60.
J. Struct. Biol., 200, 2017
|
|
6XVE
 
 | |
2BG7
 
 | | Bacillus cereus metallo-beta-lactamase (BcII) Arg (121) Cys mutant. Solved at pH4.5 using 20 Micromolar ZnSO4 in the buffer. 1mM DTT was used as a reducing agent. Cys221 is oxidized. | | Descriptor: | BETA-LACTAMASE II, GLYCEROL, SULFATE ION, ... | | Authors: | Davies, A.M, Rasia, R.M, Vila, A.J, Sutton, B.J, Fabiane, S.M. | | Deposit date: | 2004-12-17 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effect of Ph on the Active Site of an Arg121Cys Mutant of the Metallo-Beta-Lactamase from Bacillus Cereus: Implications for the Enzyme Mechanism
Biochemistry, 44, 2005
|
|
1ES2
 
 | | S96A mutant of streptomyces K15 DD-transpeptidase | | Descriptor: | DD-TRANSPEPTIDASE | | Authors: | Fonze, E, Charlier, P. | | Deposit date: | 2000-04-07 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Catalytic mechanism of the Streptomyces K15 DD-transpeptidase/penicillin-binding protein probed by site-directed mutagenesis and structural analysis.
Biochemistry, 42, 2003
|
|
7B4F
 
 | | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ). Human p53DBD-R282W mutant bound to DNA: R282W-MQ (I) | | Descriptor: | Cellular tumor antigen p53, DNA target, FORMIC ACID, ... | | Authors: | Rozenberg, H, Degtjarik, O, Shakked, Z. | | Deposit date: | 2020-12-02 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ).
Nat Commun, 12, 2021
|
|
7UNH
 
 | |
5V00
 
 | | Structure of HutD from Pseudomonas fluorescens SBW25 (Formate condition) | | Descriptor: | FORMIC ACID, GLYCEROL, Uncharacterized protein | | Authors: | Liu, Y, Johnston, J.M, Gerth, M.L, Baker, E.N, Lott, J.S, Rainey, P.B. | | Deposit date: | 2017-02-27 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a bicupin protein HutD involved in histidine utilization in Pseudomonas.
Proteins, 85, 2017
|
|
2ZGX
 
 | | Thrombin Inhibition | | Descriptor: | 1-[(2R)-2-aminobutanoyl]-N-(4-carbamimidoylbenzyl)-L-prolinamide, Hirudin variant-1, SODIUM ION, ... | | Authors: | Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-01-28 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Think twice: understanding the high potency of bis(phenyl)methane inhibitors of thrombin
J.Mol.Biol., 391, 2009
|
|
8OQD
 
 | | Dirhodium tetraacetate/ribonuclease A adduct in the P3221 space group (1 h soaking) | | Descriptor: | CHLORIDE ION, FORMIC ACID, Ribonuclease pancreatic, ... | | Authors: | Loreto, D, Merlino, A, Maity, B, Ueno, T. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Cross-Linked Crystals of Dirhodium Tetraacetate/RNase A Adduct Can Be Used as Heterogeneous Catalysts.
Inorg.Chem., 62, 2023
|
|
8OQE
 
 | | Dirhodium tetraacetate/ribonuclease A adduct in the P3221 space group (6 h soaking) | | Descriptor: | CHLORIDE ION, FORMIC ACID, Ribonuclease pancreatic, ... | | Authors: | Loreto, D, Merlino, A, Maity, B, Ueno, T. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cross-Linked Crystals of Dirhodium Tetraacetate/RNase A Adduct Can Be Used as Heterogeneous Catalysts.
Inorg.Chem., 62, 2023
|
|
8OQF
 
 | | Cross-linked crystal of Dirhodium tetraacetate/ribonuclease A adduct in the P3221 space group (low temperature data collection) | | Descriptor: | (mi2-acetato-O, O')-hexaaquo-dirhodium (II), CHLORIDE ION, ... | | Authors: | Loreto, D, Merlino, A, Maity, B, Ueno, T. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cross-Linked Crystals of Dirhodium Tetraacetate/RNase A Adduct Can Be Used as Heterogeneous Catalysts.
Inorg.Chem., 62, 2023
|
|
3ZKU
 
 | | Isopenicillin N synthase with substrate analogue AhCV | | Descriptor: | FE (III) ION, ISOPENICILLIN N SYNTHASE, N-[(5S)-5-amino-5-carboxypentanoyl]-L-homocysteyl-D-valine | | Authors: | Daruzzaman, A, Clifton, I.J, Rutledge, P.J. | | Deposit date: | 2013-01-24 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Interaction of Isopenicillin N Synthase with Homologated Substrate Analogues Delta-(L-Alpha-Aminoadipoyl)-L-Homocysteinyl-D-Xaa Characterised by Protein Crystallography.
Chembiochem, 14, 2013
|
|
3FUJ
 
 | |
5UIL
 
 | | X-ray structure of the FdtF N-formyltransferase from Salmonella enterica O60 in complex with TDP-Fuc3N and tetrahydrofolate | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, Formyltransferase, N-[4-({[(6R)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, ... | | Authors: | Woodford, C.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular architecture of an N-formyltransferase from Salmonella enterica O60.
J. Struct. Biol., 200, 2017
|
|
6Q00
 
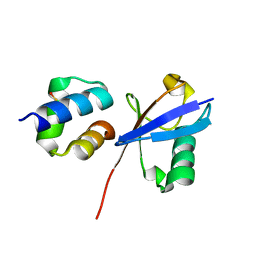 | | TDP2 UBA Domain Bound to Ubiquitin at 0.85 Angstroms Resolution, Crystal Form 1 | | Descriptor: | POTASSIUM ION, Tyrosyl-DNA phosphodiesterase 2, Ubiquitin | | Authors: | Schellenberg, M.J, Krahn, J.M, Williams, R.S. | | Deposit date: | 2019-08-01 | | Release date: | 2020-04-29 | | Last modified: | 2020-06-24 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Ubiquitin stimulated reversal of topoisomerase 2 DNA-protein crosslinks by TDP2.
Nucleic Acids Res., 48, 2020
|
|
3FTV
 
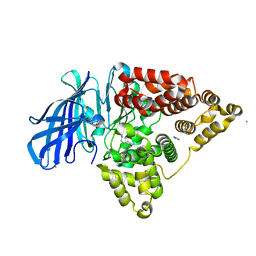 | |
1C39
 
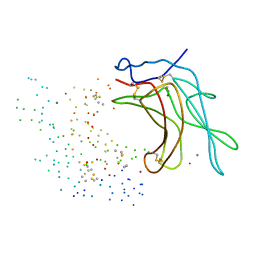 | | STRUCTURE OF CATION-DEPENDENT MANNOSE 6-PHOSPHATE RECEPTOR BOUND TO PENTAMANNOSYL PHOSPHATE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-phosphono-alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose, CATION-DEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR, ... | | Authors: | Olson, L.J, Zhang, J, Lee, Y.C, Dahms, N.M, Kim, J.J.-P. | | Deposit date: | 1999-07-25 | | Release date: | 2000-01-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for recognition of phosphorylated high mannose oligosaccharides by the cation-dependent mannose 6-phosphate receptor.
J.Biol.Chem., 274, 1999
|
|
6ZSQ
 
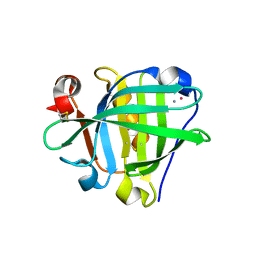 | | Crystal structure of the Cisplatin beta-Lactoglobulin adduct formed after 18 h of soaking | | Descriptor: | AMMONIA, Beta-lactoglobulin, PLATINUM (II) ION, ... | | Authors: | Balasco, N, Ferraro, G, Merlino, A. | | Deposit date: | 2020-07-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Cisplatin binding to beta-lactoglobulin: a structural study.
Dalton Trans, 49, 2020
|
|
6ZSR
 
 | | Crystal structure of the Cisplatin beta-Lactoglobulin adduct formed after 72 h of soaking | | Descriptor: | AMMONIA, Beta-lactoglobulin, PLATINUM (II) ION, ... | | Authors: | Balasco, N, Ferraro, G, Merlino, A. | | Deposit date: | 2020-07-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Cisplatin binding to beta-lactoglobulin: a structural study.
Dalton Trans, 49, 2020
|
|
1J7A
 
 | | STRUCTURE OF THE ANABAENA FERREDOXIN D68K MUTANT | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN I | | Authors: | Hurley, J.K, Weber-Main, A.M, Stankovich, M.T, Benning, M.M, Thoden, J.B, VanHooke, J.L, Holden, H.M, Chae, Y.K, Xia, B, Cheng, H, Markley, J.L, Martinez-Julvez, M, Gomez-Moreno, C, Schmeits, J.L, Tollen, G. | | Deposit date: | 2001-05-16 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants.
Biochemistry, 36, 1997
|
|
5D10
 
 | | Kinase domain of cSrc in complex with RL236 | | Descriptor: | N-[4-({4-(4-methylpiperazin-1-yl)-6-[(5-methyl-1H-pyrazol-3-yl)amino]pyrimidin-2-yl}oxy)phenyl]prop-2-enamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Becker, C, Mayer-Wrangowski, S.C, Julian, E, Rauh, D. | | Deposit date: | 2015-08-03 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Targeting Drug Resistance in EGFR with Covalent Inhibitors: A Structure-Based Design Approach.
J.Med.Chem., 58, 2015
|
|
6QMN
 
 | | Crystal structure of a Ribonuclease A-Onconase chimera | | Descriptor: | PHOSPHATE ION, Ribonuclease pancreatic | | Authors: | Esposito, L, Vitagliano, L, Ruggiero, A, Picone, D, Leone, S, Donnarumma, F. | | Deposit date: | 2019-02-07 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure, stability and aggregation propensity of a Ribonuclease A-Onconase chimera.
Int.J.Biol.Macromol., 133, 2019
|
|
