6SRA
 
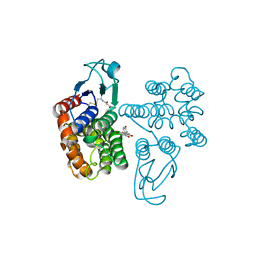 | | Crystal structure of glutathione transferase Omega 2C from Trametes versicolor in complex with naringenin | | Descriptor: | GLUTATHIONE, R-naringenin, glutathione transferase | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2019-09-05 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Diversity of Omega Glutathione Transferases in mushroom-forming fungi revealed by phylogenetic, transcriptomic, biochemical and structural approaches.
Fungal Genet Biol., 148, 2021
|
|
5OHM
 
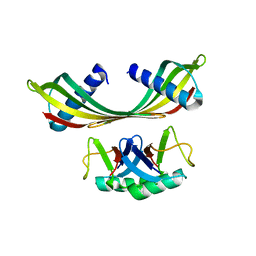 | | K33-specific affimer bound to K33 diUb | | Descriptor: | K33-specific affimer, POLYETHYLENE GLYCOL (N=34), Polyubiquitin-C | | Authors: | Michel, M.A, Komander, D. | | Deposit date: | 2017-07-17 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Ubiquitin Linkage-Specific Affimers Reveal Insights into K6-Linked Ubiquitin Signaling.
Mol. Cell, 68, 2017
|
|
6SFB
 
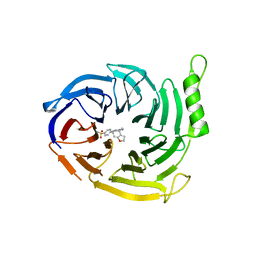 | | EED in complex with a triazolopyrimidine | | Descriptor: | GLYCEROL, N-(2,3-dihydro-1-benzofuran-4-ylmethyl)-8-(4-methylsulfonylphenyl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, Polycomb protein EED | | Authors: | Read, J.A. | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Rapid Identification of Novel Allosteric PRC2 Inhibitors.
Acs Chem.Biol., 14, 2019
|
|
6ST4
 
 | |
6STC
 
 | |
5OLG
 
 | | Structure of the A2A-StaR2-bRIL562-ZM241385 complex at 1.86A obtained from in meso soaking experiments. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, ... | | Authors: | Rucktooa, P, Cheng, R.K.Y, Segala, E, Geng, T, Errey, J.C, Brown, G.A, Cooke, R, Marshall, F.H, Dore, A.S. | | Deposit date: | 2017-07-27 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Towards high throughput GPCR crystallography: In Meso soaking of Adenosine A2A Receptor crystals.
Sci Rep, 8, 2018
|
|
5OLO
 
 | | Structure of the A2A-StaR2-bRIL562-Tozadenant complex at 3.1A obtained from in meso soaking experiments. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Rucktooa, P, Cheng, R.K.Y, Segala, E, Geng, T, Errey, J.C, Brown, G.A, Cooke, R, Marshall, F.H, Dore, A.S. | | Deposit date: | 2017-07-28 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Towards high throughput GPCR crystallography: In Meso soaking of Adenosine A2A Receptor crystals.
Sci Rep, 8, 2018
|
|
5OJP
 
 | | YCF48 bound to D1 peptide | | Descriptor: | Ycf48-like protein | | Authors: | Michoux, F, Nixon, P.J, Murray, J.W. | | Deposit date: | 2017-07-22 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Ycf48 involved in the biogenesis of the oxygen-evolving photosystem II complex is a seven-bladed beta-propeller protein.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6SGP
 
 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) - the E424M inactive mutant, in complex with a sulfamide inhibitor GluGlu | | Descriptor: | (2~{S})-2-[[(2~{S})-1,5-bis(oxidanyl)-1,5-bis(oxidanylidene)pentan-2-yl]sulfamoylamino]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Barinka, C, Shukla, S, Motlova, L. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural, Biochemical, and Computational Characterization of Sulfamides as Bimetallic Peptidase Inhibitors.
J.Chem.Inf.Model., 2024
|
|
6STH
 
 | |
5OKA
 
 | | Non-conservatively refined structure of Gan1D-E170Q, a catalytic mutant of a 6-phospho-beta-galactosidase from Geobacillus stearothermophilus | | Descriptor: | GLYCEROL, IMIDAZOLE, Putative 6-phospho-beta-galactobiosidase | | Authors: | Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2017-07-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural basis for enzyme bifunctionality - the case of Gan1D from Geobacillus stearothermophilus.
FEBS J., 284, 2017
|
|
5OLW
 
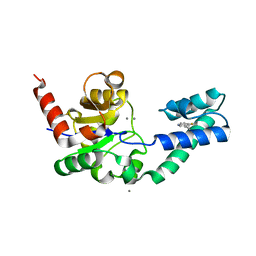 | |
6SH1
 
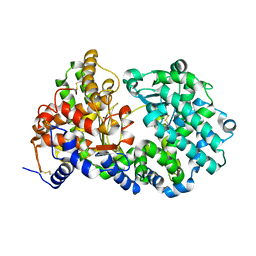 | | Crystal structure of substrate-free human neprilysin E584D. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Neprilysin, ... | | Authors: | Moss, S, Subramanian, V, Acharya, K.R. | | Deposit date: | 2019-08-05 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of peptide-bound neprilysin reveals key binding interactions.
Febs Lett., 594, 2020
|
|
5OLK
 
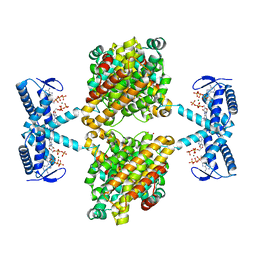 | | Crystal structure of the ATP-cone-containing NrdB from Leeuwenhoekiella blandensis | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Hasan, M, Grinberg, A.R, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2017-07-28 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Novel ATP-cone-driven allosteric regulation of ribonucleotide reductase via the radical-generating subunit.
Elife, 7, 2018
|
|
5OHU
 
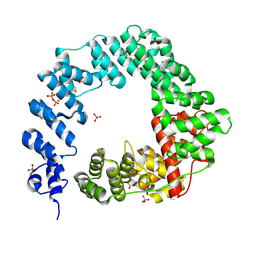 | |
5OIL
 
 | | InhA (T2A mutant) complexed with 1-cyclohexyl-3-(pyridin-3-ylmethyl)urea | | Descriptor: | 1-cyclohexyl-3-(pyridin-3-ylmethyl)urea, 2-ETHOXYETHANOL, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Convery, M.A. | | Deposit date: | 2017-07-19 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Screening of a Novel Fragment Library with Functional Complexity against Mycobacterium tuberculosis InhA.
ChemMedChem, 13, 2018
|
|
5OIS
 
 | |
6SWG
 
 | |
5OLX
 
 | |
6SWX
 
 | | Leishmania major methionyl-tRNA synthetase in complex with an allosteric inhibitor | | Descriptor: | METHIONINE, Putative methionyl-tRNA synthetase, methyl 2-[[6-[[3,4-bis(fluoranyl)phenyl]amino]-1-methyl-pyrazolo[3,4-d]pyrimidin-4-yl]amino]ethanoate | | Authors: | Robinson, D.A, Torrie, L.S, Shepherd, S.M, De Rycker, M, Thomas, M.G, Wyatt, P.G. | | Deposit date: | 2019-09-24 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of an Allosteric Binding Site in Kinetoplastid Methionyl-tRNA Synthetase.
Acs Infect Dis., 6, 2020
|
|
5OKG
 
 | | Non-conservatively refined structure of Gan1D-E170Q, a catalytic mutant of a putative 6-phospho-beta-galactosidase from Geobacillus stearothermophilus, in complex with cellobiose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, 6-O-phosphono-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2017-07-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural basis for enzyme bifunctionality - the case of Gan1D from Geobacillus stearothermophilus.
FEBS J., 284, 2017
|
|
5OLS
 
 | | Rhamnogalacturonan lyase | | Descriptor: | 4-deoxy-beta-L-threo-hex-4-enopyranuronic acid-(1-2)-alpha-L-rhamnopyranose-(1-4)-beta-D-galactopyranuronic acid, CALCIUM ION, Rhamnogalacturonan lyase, ... | | Authors: | Basle, A, Luis, A.S, Gilbert, H.J. | | Deposit date: | 2017-07-28 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides.
Nat Microbiol, 3, 2018
|
|
6SYN
 
 | | Crystal structure of Y. pestis penicillin-binding protein 3 | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2S)-2-carboxy-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, ACETATE ION, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Pankov, G, Hunter, W.N, Dawson, A. | | Deposit date: | 2019-09-30 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | The structure of penicillin-binding protein 3 from Yersinia pestis
To Be Published
|
|
6SYS
 
 | | Crystal structure of (3aR,4S,9bS)-4-(4-hydroxyphenyl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-8-sulfonamide with carbonic anhydrase 2 | | Descriptor: | (4~{S})-4-(4-hydroxyphenyl)-2,3,4,5-tetrahydro-1~{H}-cyclopenta[c]quinoline-8-sulfonamide, GLYCEROL, ZINC ION, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2019-10-01 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of (3aR,4S,9bS)-4-(4-hydroxyphenyl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-8-sulfonamide with carbonic anhydrase 2
To Be Published
|
|
6SZ5
 
 | |
