3QRY
 
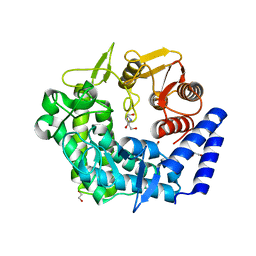 | | Analysis of a new family of widely distributed metal-independent alpha mannosidases provides unique insight into the processing of N-linked glycans, Streptococcus pneumoniae SP_2144 1-deoxymannojirimycin complex | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXYMANNOJIRIMYCIN, Putative uncharacterized protein | | Authors: | Gregg, K.J, Zandberg, W.F, Hehemann, J.-H, Whitworth, G.E, Deng, L.E, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-18 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Analysis of a New Family of Widely Distributed Metal-independent {alpha}-Mannosidases Provides Unique Insight into the Processing of N-Linked Glycans.
J.Biol.Chem., 286, 2011
|
|
4A2S
 
 | | Structure of the engineered retro-aldolase RA95.5 | | Descriptor: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE | | Authors: | Giger, L, Caner, S, Kast, P, Baker, D, Ban, N, Hilvert, D. | | Deposit date: | 2011-09-28 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evolution of a designed retro-aldolase leads to complete active site remodeling.
Nat.Chem.Biol., 9, 2013
|
|
3QPF
 
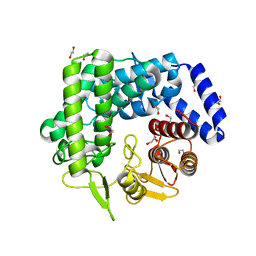 | | Analysis of a New Family of Widely Distributed Metal-independent alpha-Mannosidases Provides Unique Insight into the Processing of N-linked Glycans, Streptococcus pneumoniae SP_2144 apo-structure | | Descriptor: | 1,2-ETHANEDIOL, Putative uncharacterized protein | | Authors: | Gregg, K.J, Zandberg, W.F, Hehemann, J.-H, Whitworth, G.E, Deng, L.E, Vocadlo, D.J. | | Deposit date: | 2011-02-12 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Analysis of a New Family of Widely Distributed Metal-independent {alpha}-Mannosidases Provides Unique Insight into the Processing of N-Linked Glycans.
J.Biol.Chem., 286, 2011
|
|
3PSQ
 
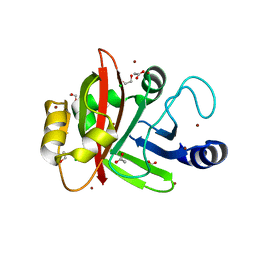 | | Crystal structure of Spy0129, a Streptococcus pyogenes class B sortase involved in pilus biogenesis | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kang, H.J, Baker, E.N. | | Deposit date: | 2010-12-02 | | Release date: | 2011-01-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure of Spy0129, a Streptococcus pyogenes class B sortase involved in pilus assembly
Plos One, 6, 2011
|
|
7B3Y
 
 | | Structure of a nanoparticle for a COVID-19 vaccine candidate | | Descriptor: | Fibronectin binding protein,2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase | | Authors: | Duyvesteyn, H.M.E, Stuart, D.I. | | Deposit date: | 2020-12-01 | | Release date: | 2021-01-13 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A COVID-19 vaccine candidate using SpyCatcher multimerization of the SARS-CoV-2 spike protein receptor-binding domain induces potent neutralising antibody responses.
Nat Commun, 12, 2021
|
|
5HEA
 
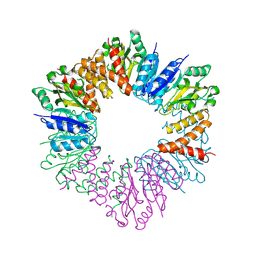 | | CgT structure in hexamer | | Descriptor: | Putative glycosyltransferase (GalT1) | | Authors: | Zhang, H, Wu, H. | | Deposit date: | 2016-01-05 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | A new helical binding domain mediates a glycosyltransferase activity of a bifunctional protein
To Be Published
|
|
5JLS
 
 | |
3FN7
 
 | |
4CNL
 
 | | Crystal structure of the Choline-binding domain of CbpL from Streptococcus pneumoniae | | Descriptor: | CHOLINE ION, GLYCEROL, PUTATIVE PNEUMOCOCCAL SURFACE PROTEIN, ... | | Authors: | Gutierrez-Fernandez, J, Bartual, S.G, Hermoso, J.A. | | Deposit date: | 2014-01-23 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Modular Architecture and Unique Teichoic Acid Recognition Features of Choline-Binding Protein L (Cbpl) Contributing to Pneumococcal Pathogenesis.
Sci.Rep., 6, 2016
|
|
3QB2
 
 | |
5IT1
 
 | | Streptomyces peucetius CYP105P2 complex with biphenyl compound | | Descriptor: | 4,4'-PROPANE-2,2-DIYLDIPHENOL, PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 | | Authors: | Lee, C.W, Lee, J.H. | | Deposit date: | 2016-03-16 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Cytochrome P450 (CYP105P2) from Streptomyces peucetius and Its Conformational Changes in Response to Substrate Binding
Int J Mol Sci, 17, 2016
|
|
3QKW
 
 | | Structure of Streptococcus parasangunini Gtf3 glycosyltransferase | | Descriptor: | Nucleotide sugar synthetase-like protein, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhu, F, Erlandsen, H, Huang, Y, Ding, L, Zhou, M, Liang, X, Ma, J.-B, Wu, H. | | Deposit date: | 2011-02-01 | | Release date: | 2011-06-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Structural and Functional Analysis of a New Subfamily of Glycosyltransferases Required for Glycosylation of Serine-rich Streptococcal Adhesins.
J.Biol.Chem., 286, 2011
|
|
5JLU
 
 | |
5XXZ
 
 | |
3G66
 
 | | The crystal structure of Streptococcus pneumoniae Sortase C provides novel insights into catalysis as well as pilin substrate specificity | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Sortase C | | Authors: | Neiers, F, Madhurantakam, C, Falker, S, Manzano, C, Dessen, A, Normark, S, Henriques-Normark, B, Achour, A. | | Deposit date: | 2009-02-06 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Two crystal structures of pneumococcal pilus sortase C provide novel insights into catalysis and substrate specificity.
J.Mol.Biol., 393, 2009
|
|
4D8I
 
 | |
5AOE
 
 | |
5AOF
 
 | |
3NBM
 
 | | The lactose-specific IIB component domain structure of the phosphoenolpyruvate:carbohydrate phosphotransferase system (PTS) from Streptococcus pneumoniae. | | Descriptor: | GLYCEROL, PTS system, lactose-specific IIBC components | | Authors: | Cuff, M.E, Chhor, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-03 | | Release date: | 2010-09-08 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of the lactose-specific IIB component domain of the phosphoenolpyruvate:carbohydrate phosphotransferase system (PTS) from Streptococcus pneumoniae.
TO BE PUBLISHED
|
|
5JUI
 
 | |
3QPB
 
 | | Crystal Structure of Streptococcus Pyogenes Uridine Phosphorylase Reveals a Subclass of the NP-I Superfamily | | Descriptor: | 1-O-phosphono-alpha-D-ribofuranose, URACIL, Uridine phosphorylase | | Authors: | Tran, T.H, Christoffersen, S, Parker, W.B, Piskur, J, Serra, I, Terreni, M, Ealick, S.E. | | Deposit date: | 2011-02-11 | | Release date: | 2011-08-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Crystal Structure of Streptococcus pyogenes Uridine Phosphorylase Reveals a Distinct Subfamily of Nucleoside Phosphorylases.
Biochemistry, 50, 2011
|
|
5I0Q
 
 | | Structure of human C4b-binding protein alpha chain CCP domains 1 and 2 in complex with the hypervariable region of mutant group A Streptococcus M2 (K65A, N66A) protein | | Descriptor: | C4b-binding protein alpha chain, M protein, serotype 2.1 | | Authors: | Buffalo, C.Z, Bahn-Suh, A.J, Ghosh, P. | | Deposit date: | 2016-02-04 | | Release date: | 2016-07-20 | | Last modified: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Conserved patterns hidden within group A Streptococcus M protein hypervariability recognize human C4b-binding protein.
Nat Microbiol, 1, 2016
|
|
4C25
 
 | | L-fuculose 1-phosphate aldolase | | Descriptor: | 1,2-ETHANEDIOL, 1,3-DIHYDROXYACETONEPHOSPHATE, GLYCEROL, ... | | Authors: | Higgins, M.A, Suits, M.D.L, Marsters, C, Boraston, A.B. | | Deposit date: | 2013-08-16 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural and Functional Analysis of Fucose-Processing Enzymes from Streptococcus Pneumoniae.
J.Mol.Biol., 426, 2014
|
|
4CUC
 
 | | Unravelling the multiple functions of the architecturally intricate Streptococcus pneumoniae beta-galactosidase, BgaA. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BETA-GALACTOSIDASE, SULFATE ION, ... | | Authors: | Singh, A.K, Pluvinage, B, Higgins, M.A, Dalia, A.B, Flynn, M, Lloyd, A.R, Weiser, J.N, Stubbs, K.A, Boraston, A.B, King, S.J. | | Deposit date: | 2014-03-17 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unravelling the Multiple Functions of the Architecturally Intricate Streptococcus Pneumoniae Beta-Galactosidase, BgaA.
Plos Pathog., 10, 2014
|
|
4D8E
 
 | |
