4OVH
 
 | | E. coli sliding clamp in complex with (R)-6-bromo-9-(2-(carboxymethylamino)-2-oxoethyl)-2,3,4,9-tetrahydro-1H-carbazole-2-carboxylic acid | | Descriptor: | (2R)-6-bromo-9-{2-[(carboxymethyl)amino]-2-oxoethyl}-2,3,4,9-tetrahydro-1H-carbazole-2-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2014-02-21 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Bacterial Sliding Clamp Inhibitors that Mimic the Sequential Binding Mechanism of Endogenous Linear Motifs.
J.Med.Chem., 58, 2015
|
|
4OVW
 
 | | ENDOGLUCANASE I COMPLEXED WITH EPOXYBUTYL CELLOBIOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(beta-D-glucopyranosyloxy)-2,2-dihydroxybutyl propanoate, ENDOGLUCANASE I | | Authors: | Davies, G.J, Schulein, M. | | Deposit date: | 1997-10-06 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the endoglucanase I from Fusarium oxysporum: native, cellobiose, and 3,4-epoxybutyl beta-D-cellobioside-inhibited forms, at 2.3 A resolution.
Biochemistry, 36, 1997
|
|
2F5P
 
 | | MutM crosslinked to undamaged DNA sampling A:T base pair IC2 | | Descriptor: | 5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*TP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*CP*G*TP*CP*CP*AP*AP*GP*TP*CP*TP*AP*CP*C)-3', ZINC ION, ... | | Authors: | Banerjee, A, Santos, W.L, Verdine, G.L. | | Deposit date: | 2005-11-26 | | Release date: | 2006-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a DNA glycosylase searching for lesions.
Science, 311, 2006
|
|
3E8Z
 
 | |
3EJT
 
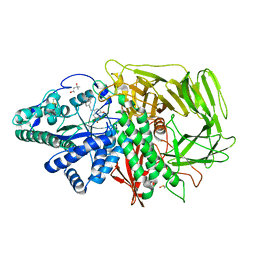 | | Golgi alpha-Mannosidase II in complex with 5-substituted swainsonine analog:(5R)-5-[2'-(4-tert-butylphenyl)ethyl]-swainsonine | | Descriptor: | (1S,2R,5R,8R,8aR)-5-[2-(4-tert-butylphenyl)ethyl]octahydroindolizine-1,2,8-triol, (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Shea, K, Rose, D.R. | | Deposit date: | 2008-09-18 | | Release date: | 2009-10-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Investigation of the Binding of 5-Substituted Swainsonine Analogues to Golgi alpha-Mannosidase II.
Chembiochem, 11, 2010
|
|
3EAW
 
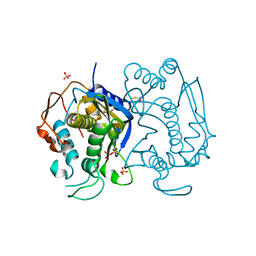 | | Replacement of Val3 in Human Thymidylate Synthase Affects Its Kinetic Properties and Intracellular Stability | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Thymidylate synthase | | Authors: | Huang, X, Gibson, L.M, Bell, B.J, Lovelace, L.L, Pena, M.M, Berger, F.G, Berger, S.H. | | Deposit date: | 2008-08-26 | | Release date: | 2010-03-02 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Replacement of Val3 in human thymidylate synthase affects its kinetic properties and intracellular stability .
Biochemistry, 49, 2010
|
|
2RFQ
 
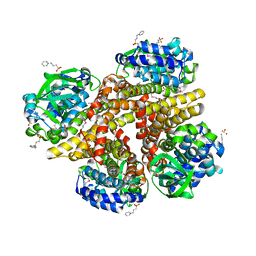 | | Crystal structure of 3-HSA hydroxylase from Rhodococcus sp. RHA1 | | Descriptor: | 3-HSA hydroxylase, oxygenase, 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE | | Authors: | Chang, C, Skarina, T, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-10-01 | | Release date: | 2007-10-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of 3-HSA hydroxylase, oxygenase from Rhodococcus sp. RHA1.
To be Published
|
|
2F7R
 
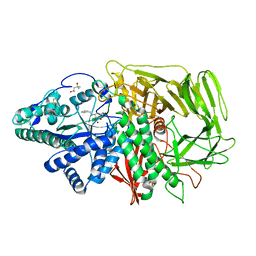 | | Golgi alpha-mannosidase II complex with benzyl-aminocyclopentitetrol | | Descriptor: | (1R,2R,3S,4S,5R)-5-(BENZYLAMINO)CYCLOPENTANE-1,2,3,4-TETROL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R. | | Deposit date: | 2005-12-01 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Golgi alpha-mannosidase II complex with benzyl-aminocyclopentitetrol
To be Published
|
|
2R6S
 
 | | Crystal structure of Gab protein | | Descriptor: | BICINE, FE (II) ION, GLYCEROL, ... | | Authors: | Lohkamp, B, Dobritzsch, D. | | Deposit date: | 2007-09-06 | | Release date: | 2008-06-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A mixture of fortunes: the curious determination of the structure of Escherichia coli BL21 Gab protein.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
5XA3
 
 | | Crystal Structure of P450BM3 with Benzyloxycarbonyl-L-prolyl-L-phenylalanine | | Descriptor: | Bifunctional cytochrome P450/NADPH-P450 reductase, DIMETHYL SULFOXIDE, PHENYLALANINE, ... | | Authors: | Shoji, O, Yanagisawa, S, Stanfield, J.K, Suzuki, K, Kasai, C, Cong, Z, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2017-03-10 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Direct Hydroxylation of Benzene to Phenol by Cytochrome P450BM3 Triggered by Amino Acid Derivatives.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6IUY
 
 | | Structure of DsGPDH of Dunaliella salina | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, GLYCEROL, Glycerol-3-phosphate dehydrogenase [NAD(+)], ... | | Authors: | He, Q, Toh, J.D, Ero, R, Qiao, Z, Kumar, V, Gao, Y.G. | | Deposit date: | 2018-12-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The unusual di-domain structure of Dunaliella salina glycerol-3-phosphate dehydrogenase enables direct conversion of dihydroxyacetone phosphate to glycerol.
Plant J., 102, 2020
|
|
6A4P
 
 | | HEWL crystals soaked in 2.5M GuHCl for 40 minutes | | Descriptor: | CHLORIDE ION, GUANIDINE, Lysozyme C, ... | | Authors: | Tushar, R, Kini, R.M, Koh, C.Y, Hosur, M.V. | | Deposit date: | 2018-06-20 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | X-ray crystallographic analysis of time-dependent binding of guanidine hydrochloride to HEWL: First steps during protein unfolding.
Int. J. Biol. Macromol., 122, 2019
|
|
3ED0
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with emodin | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, BENZAMIDINE, ... | | Authors: | Zhang, L, Zhang, H, Liu, W, Guo, Y, Shen, X, Jiang, H. | | Deposit date: | 2008-09-02 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Emodin targets the beta-hydroxyacyl-acyl carrier protein dehydratase from Helicobacter pylori: enzymatic inhibition assay with crystal structural and thermodynamic characterization
BMC MICROBIOL., 9, 2009
|
|
6E08
 
 | |
5XFM
 
 | | Crystal structure of beta-arabinopyranosidase | | Descriptor: | Alpha-glucosidase, CALCIUM ION | | Authors: | Kato, K, Okuyama, M, Yao, M. | | Deposit date: | 2017-04-10 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel glycoside hydrolase family 97 enzyme: Bifunctional beta-l-arabinopyranosidase/ alpha-galactosidase from Bacteroides thetaiotaomicron.
Biochimie, 142, 2017
|
|
3EEF
 
 | | Crystal structure of N-carbamoylsarcosine amidase from thermoplasma acidophilum | | Descriptor: | N-carbamoylsarcosine amidase related protein, ZINC ION | | Authors: | Luo, H.-B, Zheng, H, Chruszcz, M, Zimmerman, M.D, Skarina, T, Egorova, O, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-04 | | Release date: | 2008-09-16 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure and molecular modeling study of N-carbamoylsarcosine amidase Ta0454 from Thermoplasma acidophilum.
J.Struct.Biol., 169, 2010
|
|
4OWB
 
 | |
4KGX
 
 | | The R state structure of E. coli ATCase with CTP bound | | Descriptor: | Aspartate carbamoyltransferase, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Cockrell, G.M, Zheng, Y, Guo, W, Peterson, A.W, Kantrowitz, E.R. | | Deposit date: | 2013-04-29 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | New Paradigm for Allosteric Regulation of Escherichia coli Aspartate Transcarbamoylase.
Biochemistry, 52, 2013
|
|
7VDY
 
 | | Crystal structure of O-ureidoserine racemase | | Descriptor: | O-ureido-serine racemase, SULFATE ION | | Authors: | Oda, K, Matoba, Y. | | Deposit date: | 2021-09-07 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of O-ureidoserine racemase found in the d-cycloserine biosynthetic pathway.
Proteins, 90, 2022
|
|
2R8Z
 
 | | Crystal structure of YrbI phosphatase from Escherichia coli in complex with a phosphate and a calcium ion | | Descriptor: | 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase, CALCIUM ION, PHOSPHATE ION | | Authors: | Tsodikov, O.V, Aggarwal, P, Rubin, J.R, Stuckey, J.A, Woodard, R.W, Biswas, T. | | Deposit date: | 2007-09-11 | | Release date: | 2008-09-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Tail of KdsC: CONFORMATIONAL CHANGES CONTROL THE ACTIVITY OF A HALOACID DEHALOGENASE SUPERFAMILY PHOSPHATASE.
J.Biol.Chem., 284, 2009
|
|
4OWH
 
 | | PtBr6 binding to HEWL | | Descriptor: | ACETATE ION, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Tanley, S.W.M, Starkey, V.L, Lamplough, L, Kaenket, S, Helliwell, J.R. | | Deposit date: | 2014-02-02 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.484 Å) | | Cite: | The binding of platinum hexahalides (Cl, Br and I) to hen egg-white lysozyme and the chemical transformation of the PtI6 octahedral complex to a PtI3 moiety bound to His15.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4KPA
 
 | | Crystal structure of cytochrome P450 BM-3 in complex with N-palmitoylglycine (NPG) | | Descriptor: | Cytochrome P450 BM-3, N-PALMITOYLGLYCINE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Amadeo, G.A, Catalano, J, McDermott, A.E, Tong, L. | | Deposit date: | 2013-05-13 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Evidence: A Single Charged Residue Affects Substrate Binding in Cytochrome P450 BM-3.
Biochemistry, 52, 2013
|
|
6IK7
 
 | | Crystal structure of tomato beta-galactosidase (TBG) 4 in complex with beta-1,3-galactobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase, ... | | Authors: | Matsuyama, K, Nakae, S, Igarashi, K, Tada, T, Ishimaru, M. | | Deposit date: | 2018-10-15 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Substrate-recognition mechanism of tomato beta-galactosidase 4 using X-ray crystallography and docking simulation.
Planta, 252, 2020
|
|
2RBS
 
 | |
4KPR
 
 | | Tetrameric form of rat selenoprotein thioredoxin reductase 1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, SULFITE ION, ... | | Authors: | Lindqvist, Y, Sandalova, T, Xu, J, Arner, E. | | Deposit date: | 2013-05-14 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Trp114 residue of thioredoxin reductase 1 is an electron relay sensor for oxidative stress
To be Published, 2013
|
|
