4N31
 
 | |
4QR0
 
 | | Crystal structure of Streptococcus pyogenes Cas2 at pH 5.6 | | Descriptor: | CRISPR-associated endoribonuclease Cas2 | | Authors: | Bae, E, Ka, D, Kim, D. | | Deposit date: | 2014-06-30 | | Release date: | 2014-11-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of Streptococcus pyogenes Cas2 protein under different pH conditions
Biochem.Biophys.Res.Commun., 451, 2014
|
|
4HKM
 
 | |
4QR1
 
 | | Crystal structure of Streptococcus pyogenes Cas2 at pH 6.5 | | Descriptor: | CRISPR-associated endoribonuclease Cas2 | | Authors: | Bae, E, Ka, D, Kim, D. | | Deposit date: | 2014-06-30 | | Release date: | 2014-11-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Structural and functional characterization of Streptococcus pyogenes Cas2 protein under different pH conditions
Biochem.Biophys.Res.Commun., 451, 2014
|
|
3E0R
 
 | | Crystal structure of cppA protein from Streptococcus pneumoniae TIGR4 | | Descriptor: | C3-degrading proteinase (CppA protein), CHLORIDE ION | | Authors: | Nocek, B, Mulligan, R, Abdullah, J, Otwinowski, Z, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-31 | | Release date: | 2008-08-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of cppA protein from Streptococcus pneumoniae TIGR4
To be Published
|
|
4QR2
 
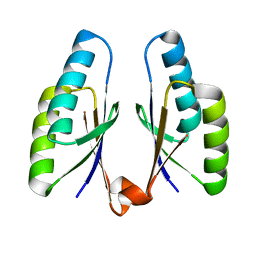 | | Crystal structure of Streptococcus pyogenes Cas2 at pH 7.5 | | Descriptor: | CRISPR-associated endoribonuclease Cas2 | | Authors: | Bae, E, Ka, D, Kim, D. | | Deposit date: | 2014-06-30 | | Release date: | 2014-11-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional characterization of Streptococcus pyogenes Cas2 protein under different pH conditions
Biochem.Biophys.Res.Commun., 451, 2014
|
|
1D2R
 
 | |
4AZC
 
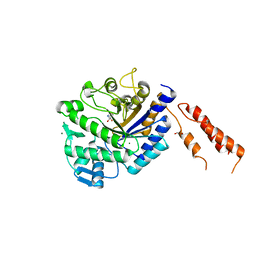 | | Differential inhibition of the tandem GH20 catalytic modules in the pneumococcal exo-beta-D-N-acetylglucosaminidase, StrH | | Descriptor: | (2S,3aR,5R,6S,7R,7aR)-5-(hydroxymethyl)-2-methyl-2,3a,5,6,7,7a-hexahydro-1H-pyrano[3,2-d][1,3]thiazole-6,7-diol, 1,2-ETHANEDIOL, BETA-N-ACETYLHEXOSAMINIDASE, ... | | Authors: | Pluvinage, B, Stubbs, K.A, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2012-06-25 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Inhibition of the Family 20 Glycoside Hydrolase Catalytic Modules in the Streptococcus Pneumoniae Exo-Beta-D-N-Acetylglucosaminidase, Strh.
Org.Biomol.Chem., 11, 2013
|
|
3O5V
 
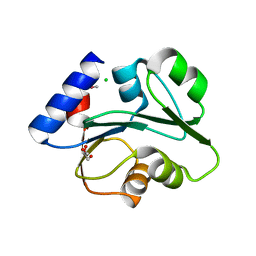 | | The Crystal Structure of the Creatinase/Prolidase N-terminal domain of an X-PRO dipeptidase from Streptococcus pyogenes to 1.85A | | Descriptor: | CHLORIDE ION, GLYCEROL, X-PRO dipeptidase | | Authors: | Stein, A.J, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-11 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of the Creatinase/Prolidase N-terminal domain of an X-PRO dipeptidase from Streptococcus pyogenes to 1.85A
To be Published
|
|
3OBH
 
 | | X-ray crystal structure of protein SP_0782 (7-79) from Streptococcus pneumoniae. Northeast Structural Genomics Consortium Target SpR104 | | Descriptor: | ACETIC ACID, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kuzin, A, Abashidze, M, Lew, S, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-08-06 | | Release date: | 2010-09-15 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | X-ray crystal structure of protein SP_0782 (7-79) from Streptococcus pneumoniae. Northeast Structural Genomics Consortium Target SpR104
To be Published
|
|
4AZ5
 
 | | Differential inhibition of the tandem GH20 catalytic modules in the pneumococcal exo-beta-D-N-acetylglucosaminidase, StrH | | Descriptor: | (2S,3aR,5R,6S,7R,7aR)-5-(hydroxymethyl)-2-methyl-2,3a,5,6,7,7a-hexahydro-1H-pyrano[3,2-d][1,3]thiazole-6,7-diol, 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Pluvinage, B, Stubbs, K.A, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2012-06-22 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Inhibition of the Family 20 Glycoside Hydrolase Catalytic Modules in the Streptococcus Pneumoniae Exo-Beta-D-N-Acetylglucosaminidase, Strh.
Org.Biomol.Chem., 11, 2013
|
|
3O92
 
 | | High resolution crystal structures of Streptococcus pneumoniae nicotinamidase with trapped intermediates provide insights into catalytic mechanism and inhibition by aldehydes | | Descriptor: | ZINC ION, nicotinamidase | | Authors: | French, J.B, Cen, Y, Sauve, A.A, Ealick, S.E. | | Deposit date: | 2010-08-03 | | Release date: | 2011-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structures of Streptococcus pneumoniae nicotinamidase with trapped intermediates provide insights into the catalytic mechanism and inhibition by aldehydes .
Biochemistry, 49, 2010
|
|
4BUG
 
 | | Pilus-presented adhesin, Spy0125 (Cpa), Cys426Ala mutant | | Descriptor: | ANCILLARY PROTEIN 1 | | Authors: | Walden, M, Crow, A, Nelson, M, Banfield, M.J. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-09 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Intramolecular Isopeptide But not Internal Thioester Bonds Confer Proteolytic and Significant Thermal Stability to the S. Pyogenes Pilus Adhesin Spy0125.
Proteins, 82, 2014
|
|
4XSA
 
 | | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis | | Descriptor: | Daunorubicin-doxorubicin polyketide synthase | | Authors: | Jackson, D.R, Valentic, T.R, Tsai, S.C, Patel, A, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
4XS7
 
 | | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis | | Descriptor: | Daunorubicin-doxorubicin polyketide synthase | | Authors: | Jackson, D.R, Valentic, T.R, Tsai, S.C, Patel, A, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
3OP1
 
 | | Crystal Structure of Macrolide-efflux Protein SP_1110 from Streptococcus pneumoniae | | Descriptor: | ACETIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Li, H, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.488 Å) | | Cite: | Crystal Structure of Macrolide-efflux Protein SP_1110 from Streptococcus pneumoniae
TO BE PUBLISHED
|
|
4C23
 
 | | L-fuculose kinase | | Descriptor: | 1,2-ETHANEDIOL, L-FUCULOSE KINASE FUCK | | Authors: | Higgins, M.A, Suits, M.D.L, Marsters, C, Boraston, A.B. | | Deposit date: | 2013-08-16 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Analysis of Fucose-Processing Enzymes from Streptococcus Pneumoniae.
J.Mol.Biol., 426, 2014
|
|
4XS9
 
 | | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis | | Descriptor: | Daunorubicin-doxorubicin polyketide synthase, N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-N-[2-(propanoylamino)ethyl]-beta-alaninamide | | Authors: | Jackson, D.R, Valentic, T.R, Tsai, S.C, Patel, A, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
4XY5
 
 | |
3P3V
 
 | |
4XSB
 
 | | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis | | Descriptor: | Daunorubicin-doxorubicin polyketide synthase | | Authors: | Jackson, D.R, Valentic, T.R, Tsai, S.C, Patel, A, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
3OVK
 
 | |
4YW4
 
 | | Streptococcus pneumoniae sialidase NanC | | Descriptor: | GLYCEROL, Neuraminidase C, PHOSPHATE ION | | Authors: | Owen, C.D, Lukacik, P, Potter, J.A, Walsh, M, Taylor, G.L. | | Deposit date: | 2015-03-20 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a novel sialidase
To be published
|
|
4C24
 
 | | L-fuculose 1-phosphate aldolase | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, L-FUCULOSE PHOSPHATE ALDOLASE, ... | | Authors: | Higgins, M.A, Suits, M.D.L, Marsters, C, Boraston, A.B. | | Deposit date: | 2013-08-16 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Analysis of Fucose-Processing Enzymes from Streptococcus Pneumoniae.
J.Mol.Biol., 426, 2014
|
|
4YW1
 
 | | Crystal Structure of Streptococcus pneumoniae NanC, complex with Neu5Ac and Neu5Ac2en following soaking with 3'SL | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Owen, C.D, Lukacik, P, Potter, J.A, Walsh, M, Taylor, G.L. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Streptococcus pneumoniae NanC: STRUCTURAL INSIGHTS INTO THE SPECIFICITY AND MECHANISM OF A SIALIDASE THAT PRODUCES A SIALIDASE INHIBITOR.
J.Biol.Chem., 290, 2015
|
|
