1MRL
 
 | | Crystal structure of streptogramin A acetyltransferase with dalfopristin | | Descriptor: | 5-(2-DIETHYLAMINO-ETHANESULFONYL)-21-HYDROXY-10-ISOPROPYL-11,19-DIMETHYL-9,26-DIOXA-3,15,28-TRIAZA-TRICYCLO[23.2.1.00,255]OCTACOSA-1(27),12,17,19,25(28)-PENTAENE-2,8,14,23-TETRAONE, Streptogramin A acetyltransferase | | Authors: | Kehoe, L.E, Snidwongse, J, Courvalin, P, Rafferty, J.B, Murray, I.A. | | Deposit date: | 2002-09-18 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Synercid (Quinupristin-Dalfopristin) Resistance in Gram-positive Bacterial Pathogens
J.Biol.Chem., 278, 2003
|
|
1VA0
 
 | | Crystal Structure of the Native Form of Uroporphyrin III C-methyl transferase from Thermus thermophilus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Uroporphyrin-III C-methyltransferase | | Authors: | Rehse, P.H, Kitao, T, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-05 | | Release date: | 2005-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of a closed-form uroporphyrinogen-III C-methyltransferase from Thermus thermophilus.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5MWW
 
 | | Sigma1.1 domain of sigmaA from Bacillus subtilis | | Descriptor: | RNA polymerase sigma factor SigA | | Authors: | Zachrdla, M, Padrta, P, Rabatinova, A, Sanderova, H, Barvik, I, Krasny, L, Zidek, L. | | Deposit date: | 2017-01-20 | | Release date: | 2017-06-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution structure of domain 1.1 of the sigma (A) factor from Bacillus subtilis is preformed for binding to the RNA polymerase core.
J. Biol. Chem., 292, 2017
|
|
5GQL
 
 | | Crystal structure of Wild Type Cypovirus Polyhedra | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Abe, S, Tabe, H, Ijiri, H, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2016-08-07 | | Release date: | 2017-02-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Engineering of Self-Assembled Porous Protein Materials in Living Cells
ACS Nano, 11, 2017
|
|
3INQ
 
 | | Crystal structure of BCL-XL in complex with W1191542 | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-(biphenyl-3-ylmethyl)piperazin-1-yl]-N-{[4-({(1R)-3-(dimethylamino)-1-[(phenylsulfanyl)methyl]propyl}amino)-3-nitrophenyl]sulfonyl}benzamide, Bcl-2-like protein 1, ... | | Authors: | Fairlie, W.D, Smith, B.J, Colman, P.M, Czabotar, P.E, Lee, E.F. | | Deposit date: | 2009-08-12 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes in Bcl-2 pro-survival proteins determine their capacity to bind ligands
J. Biol. Chem., 284, 2009
|
|
1QZ3
 
 | | CRYSTAL STRUCTURE OF MUTANT M211S/R215L OF CARBOXYLESTERASE EST2 COMPLEXED WITH HEXADECANESULFONATE | | Descriptor: | 1-HEXADECANOSULFONIC ACID, CARBOXYLESTERASE EST2 | | Authors: | De Simone, G, Mandrich, L, Menchise, V, Giordano, V, Febbraio, F, Rossi, M, Pedone, C, Manco, G. | | Deposit date: | 2003-09-15 | | Release date: | 2004-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A substrate-induced switch in the reaction mechanism of a thermophilic esterase: kinetic evidences and structural basis.
J.Biol.Chem., 279, 2004
|
|
5E7O
 
 | | Crystal structure of the perchlorate reductase PcrAB mutant W461E of PcrA from Azospira suillum PS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DMSO reductase family type II enzyme, ... | | Authors: | Tsai, C.-L, Tainer, J.A. | | Deposit date: | 2015-10-12 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Perchlorate Reductase Is Distinguished by Active Site Aromatic Gate Residues.
J.Biol.Chem., 291, 2016
|
|
2ZTB
 
 | |
6E5B
 
 | | Human Immunoproteasome 20S particle in complex with compound 1 | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Steinbacher, S, Augustin, M, Blaesse, M, Harris, S.F. | | Deposit date: | 2018-07-19 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Design and Evaluation of Highly Selective Human Immunoproteasome Inhibitors Reveal a Compensatory Process That Preserves Immune Cell Viability.
J.Med.Chem., 62, 2019
|
|
3QIY
 
 | | Crystal Structure of BoNT/A LC complexed with Hydroxamate-based Inhibitor PT-1 | | Descriptor: | 1,2-ETHANEDIOL, 4-[bis(4-chlorobenzyl)amino]-N-hydroxybutanamide, Botulinum neurotoxin type A, ... | | Authors: | Thompson, A.A, Han, G.W, Stevens, R.C. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of Three Novel Hydroxamate-Based Zinc Chelating Inhibitors of the Clostridium botulinum Serotype A Neurotoxin Light Chain Metalloprotease Reveals a Compact Binding Site Resulting from 60/70 Loop Flexibility.
Biochemistry, 50, 2011
|
|
4JT3
 
 | | Crystal Structure of TTK kinase domain with an inhibitor: 400740 | | Descriptor: | 1,2-ETHANEDIOL, 2-phenyl-N-[3-(3-sulfamoylphenyl)-2H-indazol-5-yl]acetamide, Dual specificity protein kinase TTK, ... | | Authors: | Qiu, W, Harris-Brandts, M, Feher, M, Awrey, D.E, Chirgadze, N.Y. | | Deposit date: | 2013-03-22 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of TTK kinase domain with an inhibitor: 400740
To be Published
|
|
5U5T
 
 | | Crystal structure of EED in complex with H3K27Me3 peptide and 3-(benzo[d][1,3]dioxol-4-ylmethyl)piperidine-1-carboximidamide | | Descriptor: | (3R)-3-[(2H-1,3-benzodioxol-4-yl)methyl]piperidine-1-carboximidamide, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED, ... | | Authors: | Bussiere, D, Shu, W. | | Deposit date: | 2016-12-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of EED Binders Allosterically Inhibiting the Epigenetic Polycomb Repressive Complex 2 (PRC2) Methyltransferase.
J. Med. Chem., 60, 2017
|
|
2OZW
 
 | |
5XHR
 
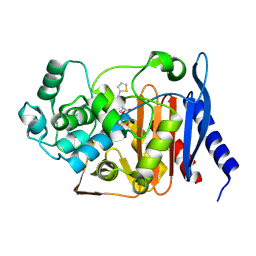 | | Crystal structure of P99 beta-lactamase in complex with a penicillin derivative MPC-1 | | Descriptor: | (2~{R},4~{S})-5,5-dimethyl-2-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-5-thiophen-2-yl-pentan-2-yl]-1,3-thiazolidine-4-carboxylic acid, Beta-lactamase | | Authors: | Pan, X, Zhao, Y. | | Deposit date: | 2017-04-24 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modified Penicillin Molecule with Carbapenem-Like Stereochemistry Specifically Inhibits Class C beta-Lactamases
Antimicrob. Agents Chemother., 61, 2017
|
|
4ZHX
 
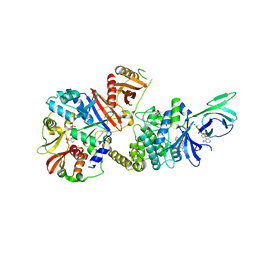 | | Novel binding site for allosteric activation of AMPK | | Descriptor: | (5S,6R,7R,9R,13cR,14R,16aS)-6-methoxy-5-methyl-7-(methylamino)-6,7,8,9,14,15,16,16a-octahydro-5H,13cH-5,9-epoxy-4b,9a,1 5-triazadibenzo[b,h]cyclonona[1,2,3,4-jkl]cyclopenta[e]-as-indacen-14-ol, 3-[4-(2-hydroxyphenyl)phenyl]-4-oxidanyl-6-oxidanylidene-7H-thieno[2,3-b]pyridine-5-carbonitrile, 5'-AMP-activated protein kinase catalytic subunit alpha-2, ... | | Authors: | Langendorf, C.G, Ngoei, K.R, Issa, S.M.A, Ling, N, Gorman, M.A, Parker, M.W, Sakamoto, K, Scott, J.W, Oakhill, J.S, Kemp, B.E. | | Deposit date: | 2015-04-27 | | Release date: | 2016-03-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis of allosteric and synergistic activation of AMPK by furan-2-phosphonic derivative C2 binding.
Nat Commun, 7, 2016
|
|
2WTJ
 
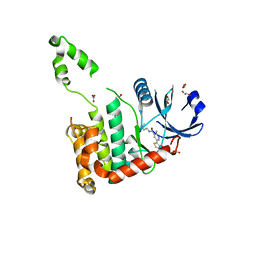 | | CRYSTAL STRUCTURE OF CHK2 IN COMPLEX WITH AN INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5-(2,3-DIHYDROTHIENO[3,4-B][1,4]DIOXIN-5-YL)-N-[2-(DIMETHYLAMINO)ETHYL]PYRIDINE-3-CARBOXAMIDE, CHECKPOINT KINASE 2, ... | | Authors: | Hilton, S, Naud, S, Caldwell, J.J, Boxall, K, Burns, S, Anderson, V.E, Antoni, L, Allen, C.E, Pearl, L.H, Oliver, A.W, Aherne, G.W, Garrett, M.D, Collins, I. | | Deposit date: | 2009-09-16 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and Characterisation of 2-Aminopyridine Inhibitors of Checkpoint Kinase 2
Bioorg.Med.Chem., 18, 2010
|
|
8WYJ
 
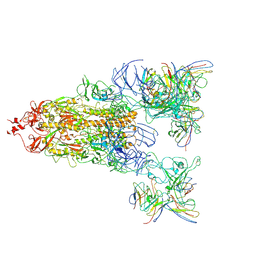 | | The global map of Omicron Subvariants Spike with two antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309-HC, S309-KC, ... | | Authors: | Yan, R.H, Wang, A.J, Yang, H.N. | | Deposit date: | 2023-10-31 | | Release date: | 2024-11-13 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the evolution and antibody evasion of SARS-CoV-2 BA.2.86 and JN.1 subvariants.
Nat Commun, 15, 2024
|
|
4ZOD
 
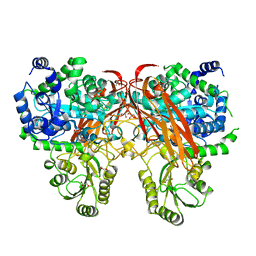 | | Crystal Structure of beta-glucosidase from Listeria innocua in complex with glucose | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Lin1840 protein, ... | | Authors: | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
7ENS
 
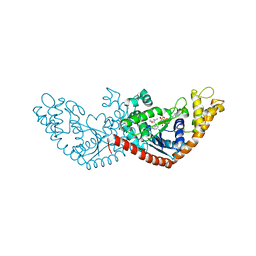 | |
2A3K
 
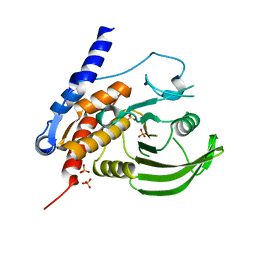 | | Crystal Structure of the Human Protein Tyrosine Phosphatase, PTPN7 (HePTP, Hematopoietic Protein Tyrosine Phosphatase) | | Descriptor: | PHOSPHATE ION, protein tyrosine phosphatase, non-receptor type 7, ... | | Authors: | Barr, A, Turnbull, A.P, Das, S, Eswaran, J, Debreczeni, J.E, Longmann, E, Smee, C, Burgess, N, Gileadi, O, Sundstrom, M, Arrowsmith, C, Edwards, A, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-24 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal structure of human receptor protein tyrosine phosphatase kappa phosphatase domain 1.
Protein Sci., 15, 2006
|
|
6TI6
 
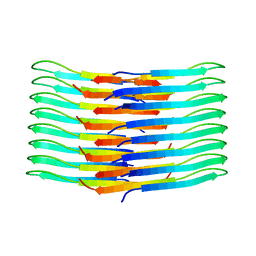 | | Mixing Abeta(1-40) and Abeta(1-42) peptides generates unique amyloid fibrils | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Cerofolini, L, Ravera, E, Bologna, S, Wiglenda, T, Boddrich, A, Purfurst, B, Benilova, A, Korsak, M, Gallo, G, Rizzo, D, Gonnelli, L, Fragai, M, De Strooper, B, Wanker, E.E, Luchinat, C. | | Deposit date: | 2019-11-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
1HOR
 
 | | STRUCTURE AND CATALYTIC MECHANISM OF GLUCOSAMINE 6-PHOSPHATE DEAMINASE FROM ESCHERICHIA COLI AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | 2-DEOXY-2-AMINO GLUCITOL-6-PHOSPHATE, GLUCOSAMINE 6-PHOSPHATE DEAMINASE, PHOSPHATE ION | | Authors: | Oliva, G, Fontes, M.R.M, Garratt, R.C, Altamirano, M.M, Calcagno, M.L, Horjales, E. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 A resolution.
Structure, 3, 1995
|
|
3ACD
 
 | | Crystal structure of hypoxanthine-guanine phosphoribosyltransferase with IMP from Thermus thermophilus HB8 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Hypoxanthine-guanine phosphoribosyltransferase, INOSINIC ACID | | Authors: | Kanagawa, M, Baba, S, Hirotsu, K, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-12-30 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structures of hypoxanthine-guanine phosphoribosyltransferase (TTHA0220) from Thermus thermophilus HB8.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
5XXN
 
 | | Crystal Structure of mutant (D286N) beta-glucosidase from Bacteroides thetaiotaomicron in complex with sophorose | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Periplasmic beta-glucosidase, ... | | Authors: | Nakajima, M, Ishiguro, R, Tanaka, N, Abe, K, Maeda, T, Miyanaga, A, Takahashi, Y, Sugimono, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-07-04 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Function and structure relationships of a beta-1,2-glucooligosaccharide-degrading beta-glucosidase.
FEBS Lett., 591, 2017
|
|
4ZLO
 
 | | Serine/threonine-protein kinase PAK1 complexed with a dibenzodiazepine: identification of an allosteric site on PAK1 | | Descriptor: | 2,8-difluoro-11-(4-methylpiperazin-1-yl)-5H-dibenzo[b,e][1,4]diazepine, GLYCEROL, Serine/threonine-protein kinase PAK 1 | | Authors: | Bellamacina, C.R, Bussiere, D.E. | | Deposit date: | 2015-05-01 | | Release date: | 2015-08-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Optimization of a Dibenzodiazepine Hit to a Potent and Selective Allosteric PAK1 Inhibitor.
Acs Med.Chem.Lett., 6, 2015
|
|
