3C6H
 
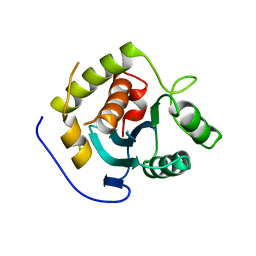 | | Crystal Structure of the RB49 gp17 nuclease domain | | Descriptor: | MAGNESIUM ION, Terminase large subunit | | Authors: | Sun, S, Rossmann, M.G. | | Deposit date: | 2008-02-04 | | Release date: | 2009-01-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces.
Cell(Cambridge,Mass.), 135, 2008
|
|
2KA4
 
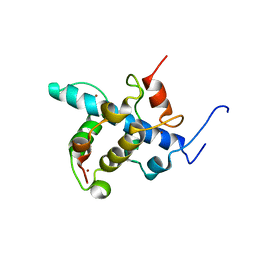 | | NMR structure of the CBP-TAZ1/STAT2-TAD complex | | Descriptor: | Crebbp protein, Signal transducer and activator of transcription 2, ZINC ION | | Authors: | Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2008-10-30 | | Release date: | 2009-04-21 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains
Embo J., 28, 2009
|
|
2KEB
 
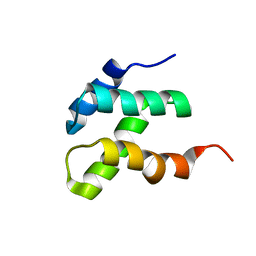 | | NMR solution structure of the N-terminal domain of the DNA polymerase alpha p68 subunit | | Descriptor: | DNA polymerase subunit alpha B | | Authors: | Huang, H, Weiner, B.E, Zhang, H, Fuller, B.E, Gao, Y, Wile, B.M, Chazin, W.J, Fanning, E. | | Deposit date: | 2009-01-28 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a DNA polymerase alpha-primase domain that docks on the SV40 helicase and activates the viral primosome.
J.Biol.Chem., 285, 2010
|
|
2XQL
 
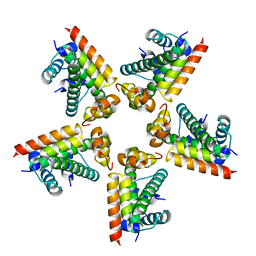 | | Fitting of the H2A-H2B histones in the electron microscopy map of the complex Nucleoplasmin:H2A-H2B histones (1:5). | | Descriptor: | HISTONE H2A-IV, HISTONE H2B 5 | | Authors: | Ramos, I, Martin-Benito, J, Finn, R, Bretana, L, Aloria, K, Arizmendi, J.M, Ausio, J, Muga, A, Valpuesta, J.M, Prado, A. | | Deposit date: | 2010-09-02 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (19.5 Å) | | Cite: | Nucleoplasmin Binds Histone H2A-H2B Dimers Through its Distal Face.
J.Biol.Chem., 285, 2010
|
|
2NNB
 
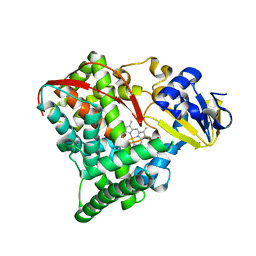 | | The Q403K mutant heme domain of flavocytochrome P450 BM3 | | Descriptor: | Bifunctional P-450:NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Clark, J.P, Anderson, J.L.R, Miles, C.S, Mowat, C.G, Reid, G.A, Chapman, S.K. | | Deposit date: | 2006-10-24 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Q403K mutant heme domain of flavocytochrome P450 BM3
To be published
|
|
2NBV
 
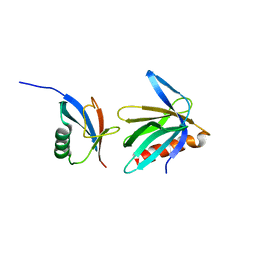 | |
2KBF
 
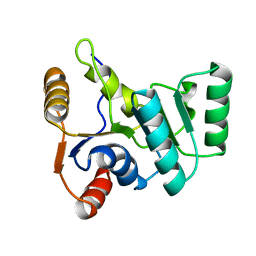 | |
2NQP
 
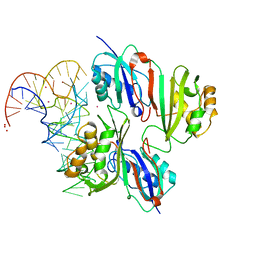 | |
2NP9
 
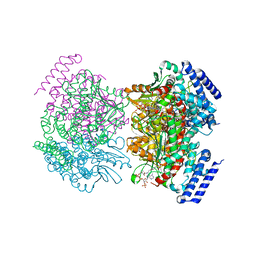 | | Crystal structure of a dioxygenase in the Crotonase superfamily | | Descriptor: | DpgC, OXYGEN MOLECULE, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-4-({3-[(2-{[(3,5-DIHYDROXYPHENYL)ACETYL]AMINO}ETHYL)AMINO]-3-OXOPROPYL}AMINO)-3-HYDROXY-2,2-DIMETHYL-4-OXOBUTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Bruner, S.D, Widboom, P.F, Fielding, E.N. | | Deposit date: | 2006-10-26 | | Release date: | 2007-05-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for cofactor-independent dioxygenation in vancomycin biosynthesis.
Nature, 447, 2007
|
|
2MTA
 
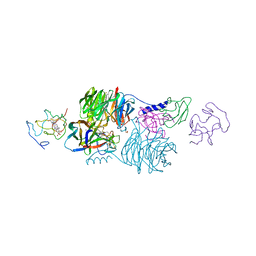 | | CRYSTAL STRUCTURE OF A TERNARY ELECTRON TRANSFER COMPLEX BETWEEN METHYLAMINE DEHYDROGENASE, AMICYANIN AND A C-TYPE CYTOCHROME | | Descriptor: | AMICYANIN, COPPER (II) ION, CYTOCHROME C551I, ... | | Authors: | Chen, L, Mathews, F.S. | | Deposit date: | 1993-10-26 | | Release date: | 1994-01-31 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of an electron transfer complex: methylamine dehydrogenase, amicyanin, and cytochrome c551i.
Science, 264, 1994
|
|
2N10
 
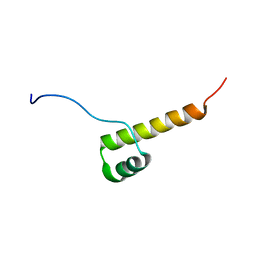 | |
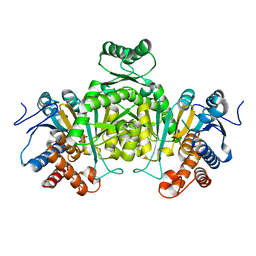
2E0C
 
 | |
2NBT
 
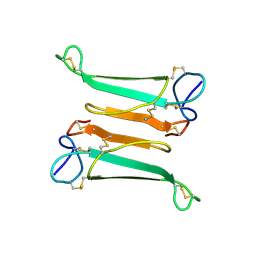 | | NEURONAL BUNGAROTOXIN, NMR, 10 STRUCTURES | | Descriptor: | NEURONAL BUNGAROTOXIN | | Authors: | Oswald, R.E, Sutcliffe, M.J, Bamberger, M, Loring, R.H, Braswell, E, Dobson, C.M. | | Deposit date: | 1997-10-29 | | Release date: | 1998-03-11 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of neuronal bungarotoxin determined by two-dimensional NMR spectroscopy: calculation of tertiary structure using systematic homologous model building, dynamical simulated annealing, and restrained molecular dynamics.
Biochemistry, 31, 1992
|
|
2NEO
 
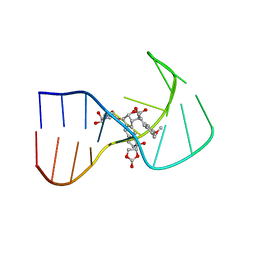 | | SOLUTION NMR STRUCTURE OF A TWO-BASE DNA BULGE COMPLEXED WITH AN ENEDIYNE CLEAVING ANALOG, 11 STRUCTURES | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*AP*TP*GP*CP*PGE*GP*CP*AP*AP*TP*TP*CP*GP*GP*G)-3'), SPIRO[[7-METHOXY-5-METHYL-1,2-DIHYDRO-NAPHTHALENE]-3,1'-[5-HYDROXY-9-[2-METHYLAMINO-2,6-DIDEOXYGALACTOPYRANOSYL-OXY]-5-(2-OXO-[1,3]DIOXOLAN-4-YL)-3A,5,9,9A-TETRAHYDRO-3H-1-OXA-CYCLOPENTA[A]-S-INDACEN-2-ONE]] | | Authors: | Stassinopoulos, A, Ji, J, Gao, X, Goldberg, I.H. | | Deposit date: | 1997-06-19 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a two-base DNA bulge complexed with an enediyne cleaving analog.
Science, 272, 1996
|
|
2E5M
 
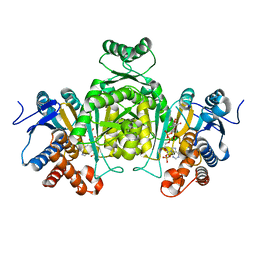 | |
2Q2C
 
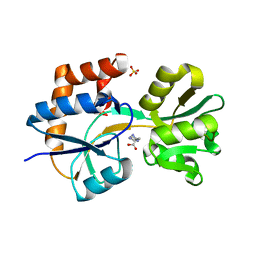 | | Crystal structures of the arginine-, lysine-, histidine-binding protein ArtJ from the thermophilic bacterium Geobacillus stearothermophilus | | Descriptor: | ArtJ, GLYCEROL, HISTIDINE, ... | | Authors: | Vahedi-Faridi, A, Scheffel, F, Eckey, V, Saenger, W, Schneider, E. | | Deposit date: | 2007-05-28 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures and mutational analysis of the arginine-, lysine-, histidine-binding protein ArtJ from Geobacillus stearothermophilus. Implications for interactions of ArtJ with its cognate ATP-binding cassette transporter, Art(MP)2
J.Mol.Biol., 375, 2008
|
|
2Q6R
 
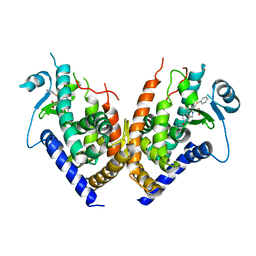 | |
2Q82
 
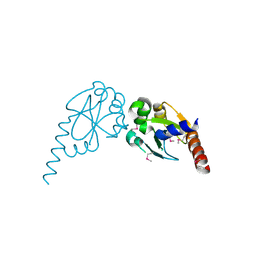 | | Crystal structure of core protein P7 from Pseudomonas phage phi12. Northeast Structural Genomics Target OC1 | | Descriptor: | Core protein P7 | | Authors: | Benach, J, Eryilmaz, E, Su, M, Seetharaman, J, Wei, H, Gottlieb, P, Hunt, J.F, Ghose, R, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-08 | | Release date: | 2007-08-07 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure and dynamics of the P7 protein from the bacteriophage phi 12.
J.Mol.Biol., 382, 2008
|
|
2Q62
 
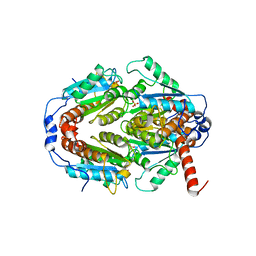 | | Crystal Structure of ArsH from Sinorhizobium meliloti | | Descriptor: | SULFATE ION, arsH | | Authors: | Ye, J, Yang, H, Bhattacharjee, H, Rosen, B.P. | | Deposit date: | 2007-06-04 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the flavoprotein ArsH from Sinorhizobium meliloti
FEBS Lett., 581, 2007
|
|
2KC8
 
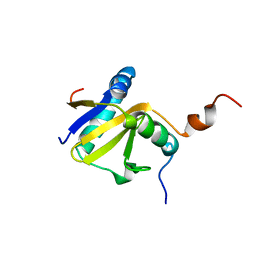 | | Structure of E. coli toxin RelE (R81A/R83A) mutant in complex with antitoxin RelBc (K47-L79) peptide | | Descriptor: | Antitoxin RelB, Toxin relE | | Authors: | Li, G, Zhang, Y, Inouye, M, Ikura, M. | | Deposit date: | 2008-12-17 | | Release date: | 2009-03-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Inhibitory mechanism of Escherichia coli RelE-RelB toxin-antitoxin module involves a helix displacement near an mRNA interferase active site.
J.Biol.Chem., 284, 2009
|
|
2PU9
 
 | |
2PUK
 
 | | Crystal structure of the binary complex between ferredoxin: thioredoxin reductase and thioredoxin m | | Descriptor: | Ferredoxin-thioredoxin reductase, catalytic chain, variable chain, ... | | Authors: | Dai, S, Friemann, R, Schurmann, P, Eklund, H. | | Deposit date: | 2007-05-09 | | Release date: | 2007-07-10 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural snapshots along the reaction pathway of ferredoxin-thioredoxin reductase.
Nature, 448, 2007
|
|
2PV0
 
 | | DNA methyltransferase 3 like protein (DNMT3L) | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, ZINC ION | | Authors: | Cheng, X. | | Deposit date: | 2007-05-09 | | Release date: | 2007-08-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA.
Nature, 448, 2007
|
|
2PVU
 
 | | Crystal structures of the arginine-, lysine-, histidine-binding protein ArtJ from the thermophilic bacterium Geobacillus stearothermophilus | | Descriptor: | ArtJ, LYSINE, SULFATE ION | | Authors: | Vahedi-Faridi, A, Scheffel, F, Eckey, V, Saenger, W, Schneider, E. | | Deposit date: | 2007-05-10 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structures and mutational analysis of the arginine-, lysine-, histidine-binding protein ArtJ from Geobacillus stearothermophilus. Implications for interactions of ArtJ with its cognate ATP-binding cassette transporter, Art(MP)2
J.Mol.Biol., 375, 2008
|
|
4F5V
 
 | | Crystal Structure of Leporine Serum Albumin | | Descriptor: | ACETATE ION, Serum albumin, TETRAETHYLENE GLYCOL, ... | | Authors: | Bujacz, A, Bujacz, G. | | Deposit date: | 2012-05-13 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structures of bovine, equine and leporine serum albumin.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
