2WF1
 
 | | Human BACE-1 in complex with 7-ethyl-N-((1S,2R)-2-hydroxy-3-(((3-(methyloxy)phenyl(methyl)amino)-1-(phenylmethyl)propyl)-1-methyl-3,4- dihydro-1H-(1,2,5)thiadiazepino(3,4,5-hi)indole-9-carboxamide 2,2- dioxide | | Descriptor: | BETA-SECRETASE 1, N-{(1S,2R)-1-benzyl-2-hydroxy-3-[(3-methoxybenzyl)amino]propyl}-7-ethyl-1-methyl-3,4-dihydro-1H-[1,2,5]thiadiazepino[3,4,5-hi]indole-9-carboxamide 2,2-dioxide | | Authors: | Charrier, N, Clarke, B, Demont, E, Dingwall, C, Dunsdon, R, Hawkins, J, Hubbard, J, Hussain, I, Maile, G, Matico, R, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Rowland, P, Soleil, V, Smith, K.J, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Wayne, G. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-19 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Second Generation of Bace-1 Inhibitors Part 2: Optimisation of the Non-Prime Side Substituent.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2WF2
 
 | | Human BACE-1 in complex with 8-ethyl-N-((1S,2R)-2-hydroxy-3-(((3-(methyloxy)phenyl)methyl)amino)-1-(phenylmethyl)propyl)-1-methyl-3,4,7, 8-tetrahydro-1H,6H-(1,2,5)thiadiazepino(5,4,3-de)quinoxaline-10- carboxamide 2,2-dioxide | | Descriptor: | BETA-SECRETASE 1, N-{(1S,2R)-1-BENZYL-2-HYDROXY-3-[(3-METHOXYBENZYL)AMINO]PROPYL}-8-ETHYL-1-METHYL-3,4,7,8-TETRAHYDRO-1H,6H-[1,2,5]THIADIAZEPINO[5,4,3-DE]QUINOXALINE-10-CARBOXAMIDE 2,2-DIOXIDE | | Authors: | Charrier, N, Clarke, B, Demont, E, Dingwall, C, Dunsdon, R, Hawkins, J, Hubbard, J, Hussain, I, Maile, G, Matico, R, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Rowland, P, Soleil, V, Smith, K.J, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Wayne, G. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-19 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Second Generation of Bace-1 Inhibitors Part 2: Optimisation of the Non-Prime Side Substituent.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2VIP
 
 | | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator | | Descriptor: | 4-(2-AMINOETHOXY)-3,5-DICHLORO-N-[3-(1-METHYLETHOXY)PHENYL]BENZAMIDE, ACETATE ION, SULFATE ION, ... | | Authors: | Frederickson, M, Callaghan, O, Chessari, G, Congreve, M, Cowan, S.R, Matthews, J.E, McMenamin, R, Smith, D, Vinkovic, M, Wallis, N.G. | | Deposit date: | 2007-12-05 | | Release date: | 2008-01-22 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator
J.Med.Chem., 51, 2008
|
|
2VIW
 
 | | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator | | Descriptor: | 4-(2-aminoethoxy)-N-(3-chloro-2-ethoxy-5-piperidin-1-ylphenyl)-3,5-dimethylbenzamide, ACETATE ION, UROKINASE-TYPE PLASMINOGEN ACTIVATOR CHAIN B | | Authors: | Frederickson, M, Callaghan, O, Chessari, G, Congreve, M, Cowan, S.R, Matthews, J.E, McMenamin, R, Smith, D, Vinkovic, M, Wallis, N.G. | | Deposit date: | 2007-12-05 | | Release date: | 2008-01-22 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator.
J.Med.Chem., 51, 2008
|
|
2VIV
 
 | | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator | | Descriptor: | 4-(2-aminoethoxy)-N-(3-chloro-5-piperidin-1-ylphenyl)-3,5-dimethylbenzamide, ACETATE ION, UROKINASE-TYPE PLASMINOGEN ACTIVATOR CHAIN B | | Authors: | Frederickson, M, Callaghan, O, Chessari, G, Congreve, M, Cowan, S.R, Matthews, J.E, McMenamin, R, Smith, D, Vinkovic, M, Wallis, N.G. | | Deposit date: | 2007-12-05 | | Release date: | 2008-01-22 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator.
J.Med.Chem., 51, 2008
|
|
2UWO
 
 | | Selective and Dual Action Orally Active Inhibitors of Thrombin and Factor Xa | | Descriptor: | (2R)-2-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}PROPENE-1-SULFONAMIDE, COAGULATION FACTOR X | | Authors: | Young, R.J, Brown, D, Burns-Kurtis, C.L, Chan, C, Convery, M.A, Hubbard, J.A, Kelly, H.A, Pateman, A.J, Patikis, A, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Zhou, P. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Selective and Dual Action Orally Active Inhibitors of Thrombin and Factor Xa.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2UWL
 
 | | Selective and Dual Action Orally Active Inhibitors of Thrombin and Factor Xa | | Descriptor: | 2-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}ETHENESULFONAMIDE, COAGULATION FACTOR X | | Authors: | Young, R.J, Brown, D, Burns-Kurtis, C.L, Chan, C, Convery, M.A, Hubbard, J.A, Kelly, H.A, Pateman, A.J, Patikis, A, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Zhou, P. | | Deposit date: | 2007-03-22 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selective and Dual Action Orally Active Inhibitors of Thrombin and Factor Xa.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2WO8
 
 | | MMP12 complex with a beta hydroxy carboxylic acid | | Descriptor: | (3S)-5-biphenyl-4-yl-3-hydroxypentanoic acid, CALCIUM ION, GLYCEROL, ... | | Authors: | Holmes, I.P, Gaines, S, Watson, S.P, Lorthioir, O, Walker, A, Baddeley, S.J, Herbert, S, Egan, D, Convery, M.A, Singh, O.M.P, Gross, J.W, Strelow, J.M, Smith, R.H, Amour, A.J, Brown, D, Martin, S.L. | | Deposit date: | 2009-07-22 | | Release date: | 2009-09-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Identification of Beta-Hydroxy Carboxylic Acids as Selective Mmp-12 Inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6WQ6
 
 | |
6WQS
 
 | | Xanthomonas citri Methionyl-tRNA synthetase in complex with REP8839 | | Descriptor: | 2-{[3-({[4-bromo-5-(1-fluoroethenyl)-3-methylthiophen-2-yl]methyl}amino)propyl]amino}quinolin-4(1H)-one, Methionine--tRNA ligase, ZINC ION | | Authors: | Mercaldi, G.F, Benedetti, C.E. | | Deposit date: | 2020-04-29 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for diaryldiamine selectivity and competition with tRNA in a type 2 methionyl-tRNA synthetase from a Gram-negative bacterium.
J.Biol.Chem., 296, 2021
|
|
6WQT
 
 | | Xanthomonas citri Methionyl-tRNA synthetase in complex with REP3123 | | Descriptor: | 5-[(3-{[(4R)-6,8-dibromo-3,4-dihydro-2H-1-benzopyran-4-yl]amino}propyl)amino]thieno[3,2-b]pyridin-7(6H)-one, Methionine--tRNA ligase, ZINC ION | | Authors: | Mercaldi, G.F, Benedetti, C.E. | | Deposit date: | 2020-04-29 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular basis for diaryldiamine selectivity and competition with tRNA in a type 2 methionyl-tRNA synthetase from a Gram-negative bacterium.
J.Biol.Chem., 296, 2021
|
|
6WQI
 
 | | Xanthomonas citri Methionyl-tRNA synthetase (apo) | | Descriptor: | Methionine--tRNA ligase, ZINC ION | | Authors: | Mercaldi, G.F, Benedetti, C.E. | | Deposit date: | 2020-04-28 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for diaryldiamine selectivity and competition with tRNA in a type 2 methionyl-tRNA synthetase from a Gram-negative bacterium.
J.Biol.Chem., 296, 2021
|
|
2WF3
 
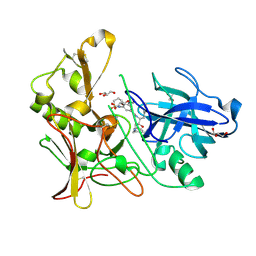 | | Human BACE-1 in complex with 6-(ethylamino)-N-((1S,2R)-2-hydroxy-3-(((3-(methyloxy)phenyl)methyl)amino)-1-(phenylmethyl)propyl)-1-methyl-1, 3,4,5-tetrahydro-2,1-benzothiazepine-8-carboxamide 2,2-dioxide | | Descriptor: | BETA-SECRETASE 1, GLYCEROL, N-{(1S,2R)-1-BENZYL-2-HYDROXY-3-[(3-METHOXYBENZYL)AMINO]PROPYL}-6-(ETHYLAMINO)-1-METHYL-1,3,4,5-TETRAHYDRO-2,1-BENZOTHIAZEPINE-8-CARBOXAMIDE 2,2-DIOXIDE | | Authors: | Charrier, N, Clarke, B, Demont, E, Dingwall, C, Dunsdon, R, Hawkins, J, Hubbard, J, Hussain, I, Maile, G, Matico, R, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Rowland, P, Soleil, V, Smith, K.J, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Wayne, G. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-19 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Second Generation of Bace-1 Inhibitors Part 2: Optimisation of the Non-Prime Side Substituent.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2WF0
 
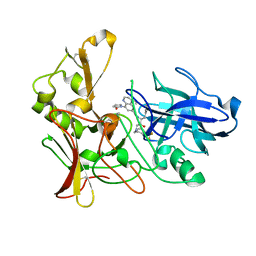 | | Human BACE-1 in complex with 4-ethyl-N-((1S,2R)-2-hydroxy-1-(phenylmethyl)-3-(((3-(trifluoromethyl)phenyl)methyl)amino)propyl)-8-(2-oxo-1-pyrrolidinyl)-6-quinolinecarboxamide | | Descriptor: | BETA-SECRETASE 1, N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{[3-(TRIFLUOROMETHYL)BENZYL]AMINO}PROPYL]-4-ETHYL-8-(2-OXOPYRROLIDIN-1-YL)QUINOLINE-6-CARBOXAMIDE | | Authors: | Charrier, N, Clarke, B, Cutler, L, Demont, E, Dingwall, C, Dunsdon, R, Hawkins, J, Howes, C, Hubbard, J, Hussain, I, Maile, G, Matico, R, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Rowland, P, Soleil, V, Smith, K.J, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Wayne, G. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-19 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Second Generation of Bace-1 Inhibitors. Part 1: The Need for Improved Pharmacokinetics.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2W26
 
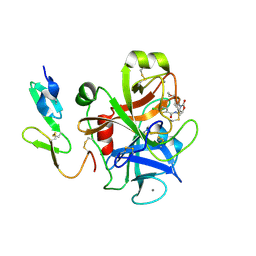 | | Factor Xa in complex with BAY59-7939 | | Descriptor: | 5-chloro-N-({(5S)-2-oxo-3-[4-(3-oxomorpholin-4-yl)phenyl]-1,3-oxazolidin-5-yl}methyl)thiophene-2-carboxamide, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION | | Authors: | Roehrig, S, Straub, A, Pohlmann, J, Lampe, T, Pernerstorfer, J, Schlemmer, K, Reinemer, P, Perzborn, E, Schaefer, M. | | Deposit date: | 2008-10-24 | | Release date: | 2008-11-11 | | Last modified: | 2021-04-28 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of the Novel Antithrombotic Agent 5-Chloro-N-({(5S)-2-Oxo-3- [4-(3-Oxomorpholin-4-Yl)Phenyl]-1,3-Oxazolidin-5-Yl}Methyl)Thiophene-2- Carboxamide (Bay 59-7939): An Oral, Direct Factor Xa Inhibitor.
J.Med.Chem., 48, 2005
|
|
2WDP
 
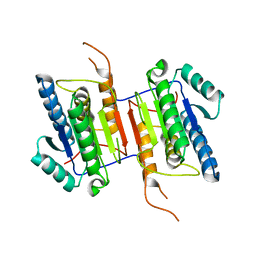 | | Crystal Structure of Ligand Free Human Caspase-6 | | Descriptor: | CASPASE-6, PHOSPHATE ION | | Authors: | Baumgartner, R, Briand, C, Meder, G, Morse, R, Renatus, M. | | Deposit date: | 2009-03-25 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of Caspase-6, a Selective Effector of Axonal Degeneration.
Biochem.J., 423, 2009
|
|
7ZVC
 
 | | Second crystal form of the mature glutamic-class prolyl-endopeptidase neprosin at 1.85 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-terminal peptidase, GLY-GLY-GLY-GLY, ... | | Authors: | Rodriguez-Banqueri, A, Eckhard, U, Del Amo-Maestro, L, Mendes, S.R, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular and in vivo studies of a glutamate-class prolyl-endopeptidase for coeliac disease therapy.
Nat Commun, 13, 2022
|
|
7ZH0
 
 | |
7ZH6
 
 | |
7ZVB
 
 | | Crystal Structure of the mature form of the glutamic-class prolyl-endopeptidase neprosin at 2.35 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-terminal peptidase, TRIETHYLENE GLYCOL, ... | | Authors: | Del Amo-Maestro, L, Eckhard, U, Rodriguez-Banqueri, A, Mendes, S.R, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular and in vivo studies of a glutamate-class prolyl-endopeptidase for coeliac disease therapy.
Nat Commun, 13, 2022
|
|
7ZHA
 
 | |
8P8G
 
 | | Nitrogenase MoFe protein from A. vinelandii beta double mutant D353G/D357G | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ... | | Authors: | Maslac, N, Wagner, T. | | Deposit date: | 2023-06-01 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Mononuclear Metal-Binding Site of Mo-Nitrogenase Is Not Required for Activity.
Jacs Au, 3, 2023
|
|
8OKL
 
 | | Crystal structure of F2F-2020185-01X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-03-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
8OKM
 
 | | Crystal structure of F2F-2020197-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-03-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
8OKK
 
 | | Crystal structure of F2F-2020184-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-03-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
