1UL2
 
 | | Solution Conformation of alpha-Conotoxin GIC | | Descriptor: | alpha-conotoxin GIC | | Authors: | Chi, S.-W, Kim, D.-H, Olivera, B.M, McIntosh, J.M, Han, K.-H. | | Deposit date: | 2003-09-06 | | Release date: | 2004-07-20 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of alpha-conotoxin GIC, a novel potent antagonist of alpha3beta2 nicotinic acetylcholine receptors
Biochem.J., 380, 2004
|
|
8AIS
 
 | | Crystal structure of cutinase PsCut from Pseudomonas saudimassiliensis | | Descriptor: | ACETATE ION, Lipase 1 | | Authors: | Zahn, M, Allen, M.D, Pickford, A.R, McGeehan, J.E. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Concentration-Dependent Inhibition of Mesophilic PETases on Poly(ethylene terephthalate) Can Be Eliminated by Enzyme Engineering.
ChemSusChem, 16, 2023
|
|
2KAP
 
 | | Solution structure of DLC1-SAM | | Descriptor: | Rho GTPase-activating protein 7 | | Authors: | Yang, S, Yang, D. | | Deposit date: | 2008-11-12 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Characterization of DLC1-SAM equilibrium unfolding at the amino acid residue level
Biochemistry, 48, 2009
|
|
2KDT
 
 | | PC1/3 DCSG sorting domain structure in DPC | | Descriptor: | Neuroendocrine convertase 1 | | Authors: | Dikeakos, J.D, Di Lello, P, Lacombe, M.J, Ghirlando, R, Legault, P, Reudelhuber, T.L, Omichinski, J.G. | | Deposit date: | 2009-01-19 | | Release date: | 2009-04-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Functional and structural characterization of a dense core secretory granule sorting domain from the PC1/3 protease.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2KJA
 
 | | Chemical shift assignments, constraints, and coordinates for CN5 scorpion toxin | | Descriptor: | Beta-toxin Cn5 | | Authors: | Prochnicka-Chalufour, A, Corzo, G, Garcia, B.I, Possani, L.D, Delepierre, M. | | Deposit date: | 2009-05-26 | | Release date: | 2009-10-06 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Cn5, a crustacean toxin found in the venom of the scorpions Centruroides noxius and Centruroides suffusus suffusus.
Biochim.Biophys.Acta, 1794, 2009
|
|
2K65
 
 | |
4V0U
 
 | | The crystal structure of ternary PP1G-PPP1R15B and G-actin complex | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, R, Yan, Y, Casado, A.C, Ron, D, Read, R.J. | | Deposit date: | 2014-09-18 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (7.88 Å) | | Cite: | G-actin provides substrate-specificity to eukaryotic initiation factor 2 alpha holophosphatases.
Elife, 4, 2015
|
|
7P1K
 
 | | Cryo EM structure of bison NHA2 in nano disc structure | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Phosphatidylinositol, mitochondrial sodium/hydrogen exchanger 9B2 | | Authors: | Matsuoka, R, Fudim, R, Jung, S, Drew, D. | | Deposit date: | 2021-07-01 | | Release date: | 2022-01-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structure, mechanism and lipid-mediated remodeling of the mammalian Na + /H + exchanger NHA2.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8DZ9
 
 | | Crystal Structure of SARS-CoV-2 Main protease G143S mutant in complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Noske, G.D, Oliva, G, Godoy, A.S. | | Deposit date: | 2022-08-06 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.664 Å) | | Cite: | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8AIR
 
 | | Crystal structure of cutinase RgCutII from Rhizobacter gummiphilus | | Descriptor: | ACETATE ION, RgCutII | | Authors: | Zahn, M, Allen, M.D, Pickford, A.R, McGeehan, J.E. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Concentration-Dependent Inhibition of Mesophilic PETases on Poly(ethylene terephthalate) Can Be Eliminated by Enzyme Engineering.
ChemSusChem, 16, 2023
|
|
2K9G
 
 | |
2K4U
 
 | | Solution structure of the SCORPION TOXIN ADWX-1 | | Descriptor: | Potassium channel toxin alpha-KTx 3.6 | | Authors: | Yin, S.J, Jiang, L, Yi, H, Han, S, Yang, D.W, Liu, M.L, Liu, H, Cao, Z.J, Wu, Y.L, Li, W.X. | | Deposit date: | 2008-06-18 | | Release date: | 2008-12-09 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Different Residues in Channel Turret Determining the Selectivity of ADWX-1 Inhibitor Peptide between Kv1.1 and Kv1.3 Channels
J.Proteome Res., 7, 2008
|
|
4QMH
 
 | | The XMAP215 family drives microtubule polymerization using a structurally diverse TOG array | | Descriptor: | LP04448p, SULFATE ION | | Authors: | Fox, J.C, Howard, A.E, Currie, J.D, Rogers, S.L, Slep, K.C. | | Deposit date: | 2014-06-16 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | The XMAP215 family drives microtubule polymerization using a structurally diverse TOG array.
Mol.Biol.Cell, 25, 2014
|
|
2KD8
 
 | |
2KDF
 
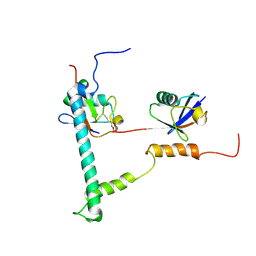 | | NMR structure of minor S5a (196-306):K48 linked diubiquitin species | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4, Ubiquitin | | Authors: | Zhang, N, Wang, Q, Ehlinger, A, Randles, L, Lary, J.W, Kang, Y, Haririnia, A, Cole, J.L, Fushman, D, Walters, K.J. | | Deposit date: | 2009-01-06 | | Release date: | 2009-09-01 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of the s5a:k48-linked diubiquitin complex and its interactions with rpn13.
Mol.Cell, 35, 2009
|
|
2KBF
 
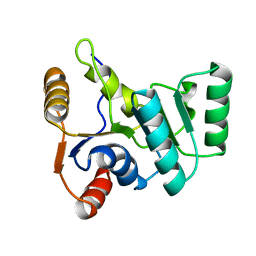 | |
4W6U
 
 | | Crystal Structure of Full-Length Split GFP Mutant E115H/T118H With Nickel Mediated Crystal Contacts, P 21 21 21 Space Group | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, NICKEL (II) ION, ... | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-20 | | Release date: | 2015-02-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
2KMO
 
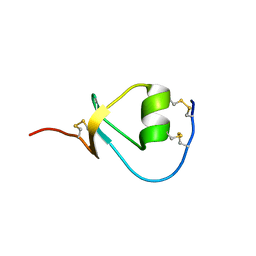 | |
2KNF
 
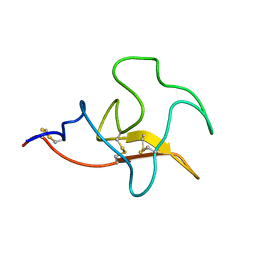 | | Solution structure and functional characterization of human plasminogen kringle 5 | | Descriptor: | Plasminogen | | Authors: | Battistel, M.D, Grishaev, A, An, S.A, Castellino, F.J, Llinas, M. | | Deposit date: | 2009-08-21 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure and functional characterization of human plasminogen kringle 5.
Biochemistry, 48, 2009
|
|
2KHC
 
 | | Bruno RRM3+ | | Descriptor: | Testis-specific RNP-type RNA binding protein | | Authors: | Lyon, A.M, Reveal, B.S, Macdonald, P.M, Hoffman, D.W. | | Deposit date: | 2009-04-01 | | Release date: | 2009-12-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Bruno protein contains an expanded RNA recognition motif.
Biochemistry, 48, 2009
|
|
2K7H
 
 | | NMR solution structure of soybean allergen Gly m 4 | | Descriptor: | Stress-induced protein SAM22 | | Authors: | Berkner, H, Neudecker, P, Mittag, D, Ballmer-Weber, B.K, Schweimer, K, Vieths, S, Roesch, P. | | Deposit date: | 2008-08-11 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Cross-reactivity of pollen and food allergens: soybean Gly m 4 is a member of the Bet v 1 superfamily and closely resembles yellow lupine proteins
Biosci.Rep., 29, 2009
|
|
2K7W
 
 | | BAX Activation is Initiated at a Novel Interaction Site | | Descriptor: | Apoptosis regulator BAX, Bcl-2-like protein 11 | | Authors: | Gavathiotis, E, Suzuki, M, Davis, M.L, Pitter, K, Bird, G.H, Katz, S.G, Tu, H.C, Kim, H, Cheng, E.H, Tjandra, N, Walensky, L.D. | | Deposit date: | 2008-08-27 | | Release date: | 2008-10-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | BAX activation is initiated at a novel interaction site.
Nature, 455, 2008
|
|
7GU5
 
 | |
2KIC
 
 | | n-NafY. N-terminal domain of NafY | | Descriptor: | Nitrogenase gamma subunit | | Authors: | Phillips, A.H, Hernandez, J.A, Erbil, K, Pelton, J.G, Wemmer, D.E, Rubio, L.M. | | Deposit date: | 2009-05-01 | | Release date: | 2010-12-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Biological activity and solution structure of the apo-dinitrogenase binding domain of NafY
J.Biol.Chem., 2010
|
|
2ERI
 
 | | Solution structure of circulin B | | Descriptor: | Circulin B | | Authors: | Craik, D.J, Daly, N.L. | | Deposit date: | 2005-10-24 | | Release date: | 2005-11-15 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Structure of circullin B and implications for antimicrobial activity of the cyclotides
INT.J.PEPT.PROTEIN RES., 11, 2005
|
|
