1XNB
 
 | |
1XNC
 
 | |
1XND
 
 | |
1XNE
 
 | | Solution Structure of Pyrococcus furiosus Protein PF0470: The Northeast Structural Genomics Consortium Target PfR14 | | Descriptor: | hypothetical protein PF0469 | | Authors: | Liu, G, Xiao, R, Parish, D, Ma, L, Sukumaran, D, Acton, T, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR data collection and analysis protocol for high-throughput protein structure determination.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1XNF
 
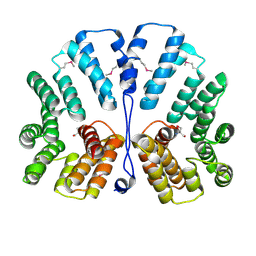 | | Crystal structure of E.coli TPR-protein NlpI | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Lipoprotein nlpI | | Authors: | Wilson, C.G, Kajander, T, Regan, L. | | Deposit date: | 2004-10-04 | | Release date: | 2004-11-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The crystal structure of NlpI. A prokaryotic tetratricopeptide repeat protein with a globular fold.
FEBS J., 272, 2005
|
|
1XNG
 
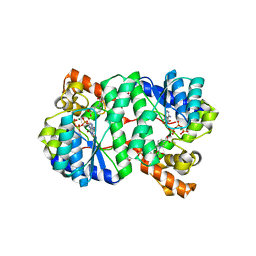 | | Crystal Structure of NH3-dependent NAD+ synthetase from Helicobacter pylori | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NH(3)-dependent NAD(+) synthetase, ... | | Authors: | Kang, G.B, Kim, Y.S, Im, Y.J, Rho, S.H, Lee, J.H, Eom, S.H. | | Deposit date: | 2004-10-05 | | Release date: | 2005-04-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of NH3-dependent NAD+ synthetase from Helicobacter pylori
Proteins, 58, 2005
|
|
1XNH
 
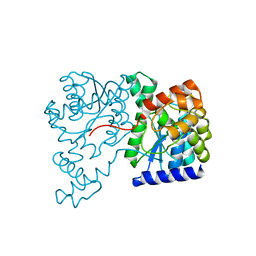 | | Crystal Structure of NH3-dependent NAD+ synthetase from Helicobacter pylori | | Descriptor: | NH(3)-dependent NAD(+) synthetase | | Authors: | Kang, G.B, Kim, Y.S, Im, Y.J, Rho, S.H, Lee, J.H, Eom, S.H. | | Deposit date: | 2004-10-05 | | Release date: | 2005-04-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of NH3-dependent NAD+ synthetase from Helicobacter pylori
Proteins, 58, 2005
|
|
1XNI
 
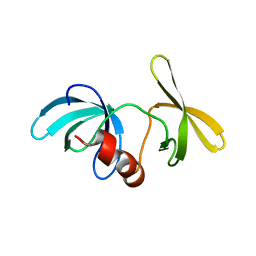 | | Tandem Tudor Domain of 53BP1 | | Descriptor: | Tumor suppressor p53-binding protein 1 | | Authors: | Huyen, Y, Zgheib, O, DiTullio Jr, R.A, Gorgoulis, V.G, Zacharatos, P, Petty, T.J, Sheston, E.A, Mellert, H.S, Stavridi, E.S, Halazonetis, T.D. | | Deposit date: | 2004-10-05 | | Release date: | 2004-11-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks
Nature, 432, 2004
|
|
1XNJ
 
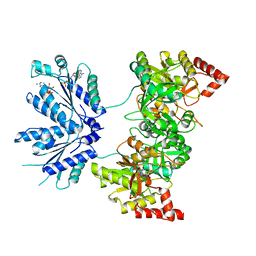 | | APS complex of human PAPS synthetase 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-PHOSPHOSULFATE, Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthetase 1 | | Authors: | Harjes, S, Bayer, P, Scheidig, A.J. | | Deposit date: | 2004-10-05 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The crystal structure of human PAPS synthetase 1 reveals asymmetry in substrate binding
J.Mol.Biol., 347, 2005
|
|
1XNK
 
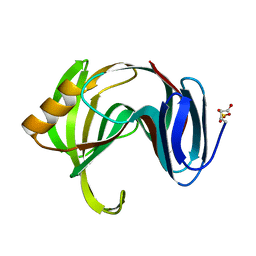 | | Beta-1,4-xylanase from Chaetomium thermophilum complexed with methyl thioxylopentoside | | Descriptor: | 4-thio-beta-D-xylopyranose-(1-4)-4-thio-beta-D-xylopyranose-(1-4)-methyl 4-thio-alpha-D-xylopyranoside, SULFATE ION, endoxylanase 11A | | Authors: | Hakanpaa, J, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2004-10-05 | | Release date: | 2005-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Determination of thioxylo-oligosaccharide binding to family 11 xylanases using electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry and X-ray crystallography
FEBS J., 272, 2005
|
|
1XNL
 
 | | ASLV fusion peptide | | Descriptor: | membrane protein gp37 | | Authors: | Cheng, S.F, Wu, C.W, Kantchev, E.A, Chang, D.K. | | Deposit date: | 2004-10-05 | | Release date: | 2005-01-11 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure and membrane interaction of the internal fusion peptide of avian sarcoma leukosis virus
Eur.J.Biochem., 271, 2004
|
|
1XNN
 
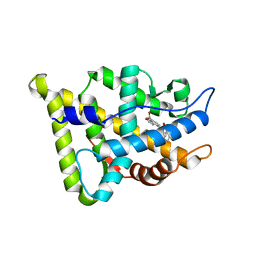 | | CRYSTAL STRUCTURE OF THE RAT ANDROGEN RECEPTOR LIGAND BINDING DOMAIN T877A MUTANT COMPLEX WITH (3A-ALPHA-,4-ALPHA 7-ALPHA-,7A-ALPHA-)-3A,4,7,7A-TETRAHYDRO-2-(4-NITRO-1-NAPHTHALENYL)-4,7-ETHANO-1H-ISOINDOLE-1,3(2H)-DIONE. | | Descriptor: | Androgen receptor, REL-(3AR,4S,7R,7AS)-3A,4,7,7A-TETRAHYDRO-2-(4-NITRO-1-NAPHTHALENYL)-4,7-ETHANO-1H-ISOINDOLE-1,3(2H)-DIONE | | Authors: | Sack, J. | | Deposit date: | 2004-10-05 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure based approach to the design of bicyclic-1H-isoindole-1,3(2H)-dione based androgen receptor antagonists.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1XNQ
 
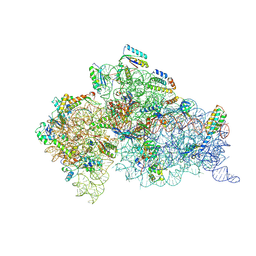 | |
1XNR
 
 | |
1XNS
 
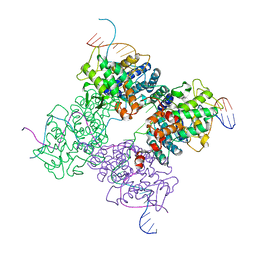 | | Peptide trapped Holliday junction intermediate in Cre-loxP recombination | | Descriptor: | Recombinase CRE, loxP DNA | | Authors: | Ghosh, K, Lau, C.K, Guo, F, Segall, A.M, Van Duyne, G.D. | | Deposit date: | 2004-10-05 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Peptide trapping of the Holliday junction intermediate in Cre-loxP site-specific recombination.
J.Biol.Chem., 280, 2005
|
|
1XNT
 
 | |
1XNV
 
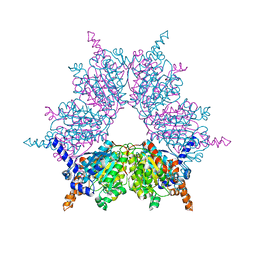 | | Acyl-CoA Carboxylase Beta Subunit from S. coelicolor (PccB), apo form #1 | | Descriptor: | propionyl-CoA carboxylase complex B subunit | | Authors: | Diacovich, L, Mitchell, D.L, Pham, H, Gago, G, Melgar, M.M, Khosla, C, Gramajo, H, Tsai, S.-C. | | Deposit date: | 2004-10-05 | | Release date: | 2004-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the beta-Subunit of Acyl-CoA Carboxylase: Structure-Based Engineering of Substrate Specificity
Biochemistry, 43, 2004
|
|
1XNW
 
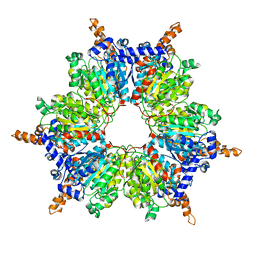 | | Acyl-CoA Carboxylase Beta Subunit from S. coelicolor (PccB), apo form #2, mutant D422I | | Descriptor: | propionyl-CoA carboxylase complex B subunit | | Authors: | Diacovich, L, Mitchell, D.L, Pham, H, Gago, G, Melgar, M.M, Khosla, C, Gramajo, H, Tsai, S.-C. | | Deposit date: | 2004-10-05 | | Release date: | 2004-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the beta-Subunit of Acyl-CoA Carboxylase: Structure-Based Engineering of Substrate Specificity
Biochemistry, 43, 2004
|
|
1XNX
 
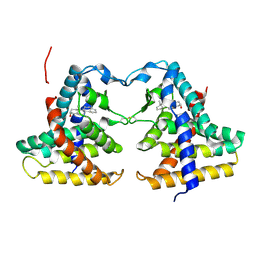 | | Crystal structure of constitutive androstane receptor | | Descriptor: | 16,17-ANDROSTENE-3-OL, constitutive androstane receptor | | Authors: | Fernandez, E. | | Deposit date: | 2004-10-05 | | Release date: | 2005-01-04 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the murine constitutive androstane receptor complexed to androstenol; a molecular basis for inverse agonism
Mol.Cell, 16, 2004
|
|
1XNY
 
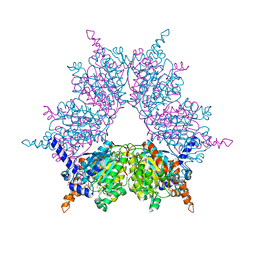 | | Biotin and propionyl-CoA bound to Acyl-CoA Carboxylase Beta Subunit from S. coelicolor (PccB) | | Descriptor: | BIOTIN, propionyl Coenzyme A, propionyl-CoA carboxylase complex B subunit | | Authors: | Diacovich, L, Mitchell, D.L, Pham, H, Gago, G, Melgar, M.M, Khosla, C, Gramajo, H, Tsai, S.-C. | | Deposit date: | 2004-10-05 | | Release date: | 2004-11-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the beta-Subunit of Acyl-CoA Carboxylase: Structure-Based Engineering of Substrate Specificity
Biochemistry, 43, 2004
|
|
1XNZ
 
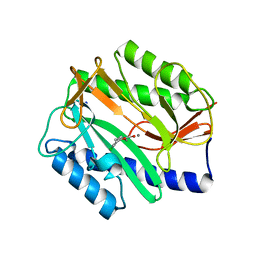 | | Crystal Structure of Mn(II) form of E. coli. Methionine Aminopeptidase in complex with 5-(2-chlorophenyl)furan-2-carboxylic acid | | Descriptor: | 5-(2-CHLOROPHENYL)FURAN-2-CARBOXYLIC ACID, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.-Z, Xie, S.-X, Huang, M, Huang, W.-J, Lu, J.-P, Ma, Z.-Q. | | Deposit date: | 2004-10-05 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Metalloform-Selective Inhibitors of Escherichia coli Methionine Aminopeptidase and X-ray Structure of a Mn(II)-Form Enzyme Complexed with an Inhibitor.
J.Am.Chem.Soc., 126, 2004
|
|
1XO0
 
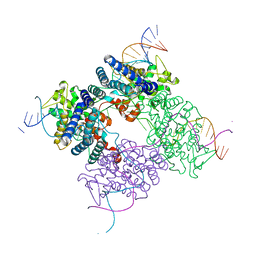 | | High resolution structure of the holliday junction intermediate in cre-loxp site-specific recombination | | Descriptor: | Recombinase CRE, loxP | | Authors: | Ghosh, K, Lau, C.K, Guo, F, Segall, A.M, Van Duyne, G.D. | | Deposit date: | 2004-10-05 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Peptide trapping of the Holliday junction intermediate in Cre-loxP site-specific recombination.
J.Biol.Chem., 280, 2005
|
|
1XO1
 
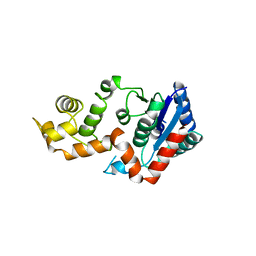 | | T5 5'-EXONUCLEASE MUTANT K83A | | Descriptor: | 5'-EXONUCLEASE | | Authors: | Ceska, T.A, Suck, D, Sayers, J.R. | | Deposit date: | 1998-11-19 | | Release date: | 1999-04-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis of conserved lysine residues in bacteriophage T5 5'-3' exonuclease suggests separate mechanisms of endo-and exonucleolytic cleavage.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1XO2
 
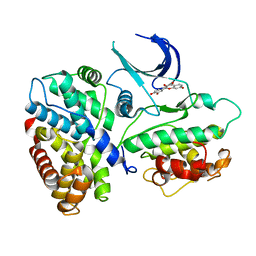 | | Crystal structure of a human cyclin-dependent kinase 6 complex with a flavonol inhibitor, fisetin | | Descriptor: | 3,7,3',4'-TETRAHYDROXYFLAVONE, Cell division protein kinase 6, Cyclin | | Authors: | Lu, H.S, Chang, D.J, Baratte, B, Meijer, L, Schulze-Gahmen, U. | | Deposit date: | 2004-10-05 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of a Human Cyclin-Dependent Kinase 6 Complex with a Flavonol Inhibitor, Fisetin.
J.Med.Chem., 48, 2005
|
|
1XO3
 
 | | Solution Structure of Ubiquitin like protein from Mus Musculus | | Descriptor: | RIKEN cDNA 2900073H19 | | Authors: | Singh, S, Tonelli, M, Tyler, R.C, Bahrami, A, Lee, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-10-05 | | Release date: | 2004-10-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the AAH26994.1 protein from Mus musculus, a putative eukaryotic Urm1.
Protein Sci., 14, 2005
|
|
