2C1V
 
 | |
1BT7
 
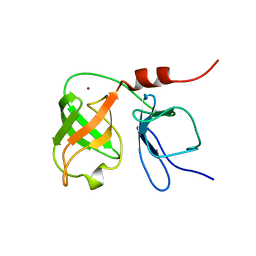 | | THE SOLUTION NMR STRUCTURE OF THE N-TERMINAL PROTEASE DOMAIN OF THE HEPATITIS C VIRUS (HCV) NS3-PROTEIN, FROM BK STRAIN, 20 STRUCTURES | | Descriptor: | NS3 SERINE PROTEASE, ZINC ION | | Authors: | Barbato, G, Cicero, D.O, Nardi, M.C, Steinkuhler, C, Cortese, R, De Francesco, R, Bazzo, R. | | Deposit date: | 1998-09-01 | | Release date: | 1999-06-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal proteinase domain of the hepatitis C virus (HCV) NS3 protein provides new insights into its activation and catalytic mechanism.
J.Mol.Biol., 289, 1999
|
|
2HM8
 
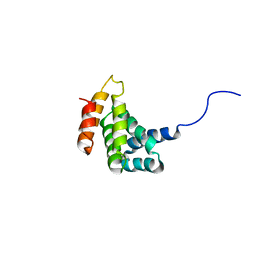 | | Solution Structure of the C-terminal MA-3 domain of Pdcd4 | | Descriptor: | Pdcd4 C-terminal MA-3 domain | | Authors: | Waters, L.C, Veverka, V, Bohm, M, Muskett, F.W, Choong, P.T, Klempnauer, K.H, Carr, M.D. | | Deposit date: | 2006-07-11 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal MA-3 domain of the tumour suppressor protein Pdcd4 and characterization of its interaction with eIF4A
Oncogene, 26, 2007
|
|
4HS8
 
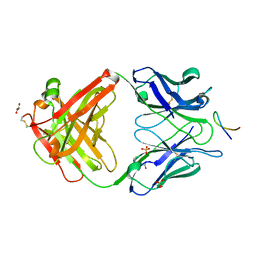 | |
1MZ4
 
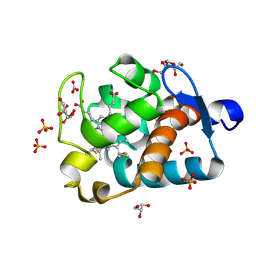 | | Crystal Structure of Cytochrome c550 from Thermosynechococcus elongatus | | Descriptor: | BICARBONATE ION, GLYCEROL, HEME C, ... | | Authors: | Kerfeld, C.A, Sawaya, M.R, Bottin, H, Tran, K.T, Sugiura, M, Kirilovsky, D, Krogmann, D, Yeates, T.O, Boussac, A. | | Deposit date: | 2002-10-05 | | Release date: | 2003-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and EPR characterization of the soluble form of cytochrome c-550 and of the psbV2 gene product from the cyanobacterium Thermosynechococcus elongatus.
Plant Cell.Physiol., 44, 2003
|
|
3I01
 
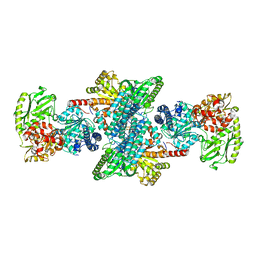 | | Native structure of bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase from Moorella thermoacetica, water-bound C-cluster. | | Descriptor: | ACETATE ION, COPPER (I) ION, Carbon monoxide dehydrogenase/acetyl-CoA synthase subunit alpha, ... | | Authors: | Kung, Y, Doukov, T.I, Drennan, C.L. | | Deposit date: | 2009-06-24 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic snapshots of cyanide- and water-bound C-clusters from bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase.
Biochemistry, 48, 2009
|
|
2L3L
 
 | | The solution structure of the N-terminal domain of human Tubulin Binding Cofactor C reveals a platform for the interaction with ab-tubulin | | Descriptor: | Tubulin-specific chaperone C | | Authors: | Garcia-Mayoral, M.F, Castano, R, Lopez-Fanarraga, M.L, Zabala, J.C, Rico, M, Bruix, M. | | Deposit date: | 2010-09-14 | | Release date: | 2011-09-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of human tubulin binding cofactor C reveals a platform for tubulin interaction
To be Published
|
|
1DTL
 
 | | CRYSTAL STRUCTURE OF CALCIUM-SATURATED (3CA2+) CARDIAC TROPONIN C COMPLEXED WITH THE CALCIUM SENSITIZER BEPRIDIL AT 2.15 A RESOLUTION | | Descriptor: | 1-ISOBUTOXY-2-PYRROLIDINO-3[N-BENZYLANILINO] PROPANE, CALCIUM ION, CARDIAC TROPONIN C | | Authors: | Li, Y, Love, M.L, Putkey, J.A, Cohen, C. | | Deposit date: | 2000-01-12 | | Release date: | 2000-05-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Bepridil opens the regulatory N-terminal lobe of cardiac troponin C.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
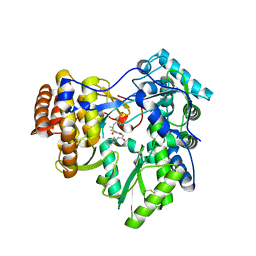
3LKH
 
 | |
4DEI
 
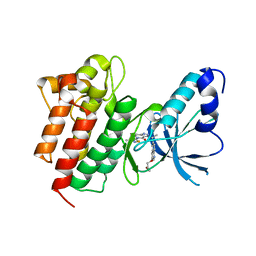 | | Crystal structure of c-Met in complex with triazolopyridinone inhibitor 24 | | Descriptor: | 3-{(1S)-1-[3-(2-methoxyethoxy)quinolin-6-yl]ethyl}-5-(3-methyl-1,2-thiazol-5-yl)-3,5-dihydro-4H-[1,2,3]triazolo[4,5-c]pyridin-4-one, Hepatocyte growth factor receptor | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2012-01-20 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery and optimization of a potent and selective triazolopyridinone series of c-Met inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1R5Z
 
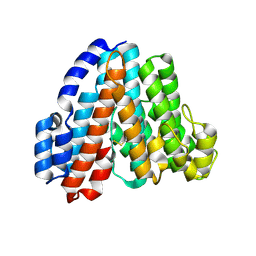 | | Crystal Structure of Subunit C of V-ATPase | | Descriptor: | V-type ATP synthase subunit C | | Authors: | Iwata, M, Imamura, H, Stambouli, E, Ikeda, C, Tamakoshi, M, Nagata, K, Makyio, H, Hankamer, B, Barber, J, Yoshida, M, Yokoyama, K, Iwata, S. | | Deposit date: | 2003-10-14 | | Release date: | 2004-01-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a central stalk subunit C and reversible association/dissociation of vacuole-type ATPase.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
3GEF
 
 | | Crystal structure of the R482W mutant of lamin A/C | | Descriptor: | Lamin-A/C | | Authors: | Magracheva, E, Kozlov, S, Stuart, C, Wlodawer, A, Zdanov, A. | | Deposit date: | 2009-02-25 | | Release date: | 2009-08-04 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the lamin A/C R482W mutant responsible for dominant familial partial lipodystrophy (FPLD).
Acta Crystallogr.,Sect.F, 65, 2009
|
|
1W2N
 
 | | Deacetoxycephalosporin C synthase (with a N-terminal his-tag) in complex with Fe(II) and ampicillin | | Descriptor: | (2S,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, DEACETOXYCEPHALOSPORIN C SYNTHASE, FE (III) ION | | Authors: | Oster, L.M, Terwisscha Van Scheltinga, A.C, Valegard, K, Mackenzie Hose, A, Dubus, A, Hajdu, J, Andersson, I. | | Deposit date: | 2004-07-07 | | Release date: | 2004-09-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational Flexibility of the C Terminus with Implications for Substrate Binding and Catalysis Revealed in a New Crystal Form of Deacetoxycephalosporin C Synthase
J.Mol.Biol., 343, 2004
|
|
3I04
 
 | | Cyanide-bound structure of bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase from Moorella thermoacetica, cyanide-bound C-cluster | | Descriptor: | ACETATE ION, COPPER (I) ION, CYANIDE ION, ... | | Authors: | Kung, Y, Doukov, T.I, Drennan, C.L. | | Deposit date: | 2009-06-24 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic snapshots of cyanide- and water-bound C-clusters from bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase.
Biochemistry, 48, 2009
|
|
4DEH
 
 | | Crystal structure of c-Met in complex with triazolopyridinone inhibitor 3 | | Descriptor: | 5-phenyl-3-(quinolin-6-ylmethyl)-3,5,6,7-tetrahydro-4H-[1,2,3]triazolo[4,5-c]pyridin-4-one, Hepatocyte growth factor receptor | | Authors: | Whittington, D.A, Bellon, S.F, Long, A.M. | | Deposit date: | 2012-01-20 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and optimization of a potent and selective triazolopyridinone series of c-Met inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1BLQ
 
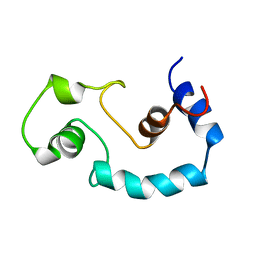 | | STRUCTURE AND INTERACTION SITE OF THE REGULATORY DOMAIN OF TROPONIN-C WHEN COMPLEXED WITH THE 96-148 REGION OF TROPONIN-I, NMR, 29 STRUCTURES | | Descriptor: | N-TROPONIN C | | Authors: | Mckay, R.T, Pearlstone, J.R, Corson, D.C, Gagne, S.M, Smillie, L.B, Sykes, B.D. | | Deposit date: | 1998-07-19 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and interaction site of the regulatory domain of troponin-C when complexed with the 96-148 region of troponin-I.
Biochemistry, 37, 1998
|
|
2TEC
 
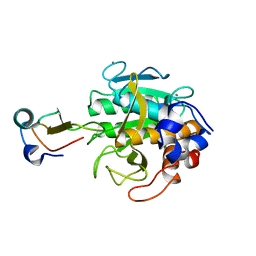 | | MOLECULAR DYNAMICS REFINEMENT OF A THERMITASE-EGLIN-C COMPLEX AT 1.98 ANGSTROMS RESOLUTION AND COMPARISON OF TWO CRYSTAL FORMS THAT DIFFER IN CALCIUM CONTENT | | Descriptor: | CALCIUM ION, EGLIN C, THERMITASE | | Authors: | Gros, P, Betzel, C, Dauter, Z, Wilson, K.S, Hol, W.G.J. | | Deposit date: | 1990-10-26 | | Release date: | 1992-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Molecular dynamics refinement of a thermitase-eglin-c complex at 1.98 A resolution and comparison of two crystal forms that differ in calcium content.
J.Mol.Biol., 210, 1989
|
|
1P5X
 
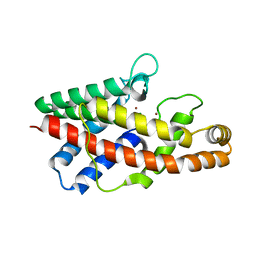 | | STRUCTURE OF THE D55N MUTANT OF PHOSPHOLIPASE C FROM BACILLUS CEREUS | | Descriptor: | Phospholipase C, ZINC ION | | Authors: | Antikainen, N.M, Monzingo, A.F, Franklin, C.L, Robertus, J.D, Martin, S.F. | | Deposit date: | 2003-04-28 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Using X-ray crystallography of the Asp55Asn mutant of the phosphatidylcholine-preferring phospholipase C from Bacillus cereus to support the mechanistic role of Asp55 as the general base.
Arch.Biochem.Biophys., 417, 2003
|
|
1P6E
 
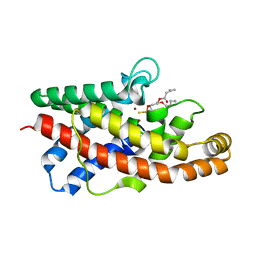 | | STRUCTURE OF THE D55N MUTANT OF PHOSPHOLIPASE C FROM BACILLUS CEREUS IN COMPLEX WITH 1,2-DI-N-PENTANOYL-SN-GLYCERO-3-DITHIOPHOSPHOCHOLINE | | Descriptor: | 1,2-DI-N-PENTANOYL-SN-GLYCERO-3-DITHIOPHOSPHOCHOLINE, Phospholipase C, ZINC ION | | Authors: | Antikainen, N.M, Monzingo, A.F, Franklin, C.L, Robertus, J.D, Martin, S.F. | | Deposit date: | 2003-04-29 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Using X-ray crystallography of the Asp55Asn mutant of the phosphatidylcholine-preferring phospholipase C from Bacillus cereus to support the mechanistic role of Asp55 as the general base.
Arch.Biochem.Biophys., 417, 2003
|
|
2A4J
 
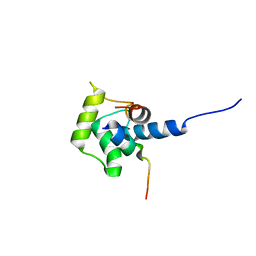 | | Solution structure of the C-terminal domain (T94-Y172) of the human centrin 2 in complex with a 17 residues peptide (P1-XPC) from xeroderma pigmentosum group C protein | | Descriptor: | 17-mer peptide P1-XPC from DNA-repair protein complementing XP-C cells, Centrin 2 | | Authors: | Yang, A, Miron, S, Mouawad, L, Duchambon, P, Blouquit, Y, Craescu, C.T. | | Deposit date: | 2005-06-29 | | Release date: | 2005-07-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Flexibility and plasticity of human centrin 2 binding to the xeroderma pigmentosum group C protein (XPC) from nuclear excision repair.
Biochemistry, 45, 2006
|
|
1P6D
 
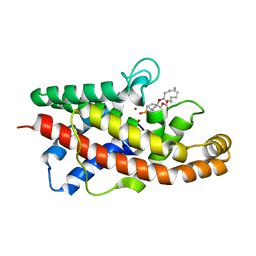 | | STRUCTURE OF THE D55N MUTANT OF PHOSPHOLIPASE C FROM BACILLUS CEREUS IN COMPLEX WITH (3S)-3,4,DI-N-HEXANOYLOXYBUTYL-1-PHOSPHOCHOLINE | | Descriptor: | (3S)-3,4-DI-N-HEXANOYLOXYBUTYL-1-PHOSPHOCHOLINE, PHOSPHOLIPASE C, ZINC ION | | Authors: | Antikainen, N.M, Monzingo, A.F, Franklin, C.L, Robertus, J.D, Martin, S.F. | | Deposit date: | 2003-04-29 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Using X-ray crystallography of the Asp55Asn mutant of the phosphatidylcholine-preferring phospholipase C from Bacillus cereus to support the mechanistic role of Asp55 as the general base.
Arch.Biochem.Biophys., 417, 2003
|
|
1ELU
 
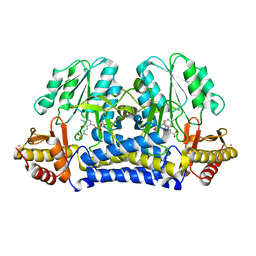 | | COMPLEX BETWEEN THE CYSTINE C-S LYASE C-DES AND ITS REACTION PRODUCT CYSTEINE PERSULFIDE. | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-PROPIONIC ACID, L-CYSTEINE/L-CYSTINE C-S LYASE, POTASSIUM ION, ... | | Authors: | Clausen, T, Kaiser, J.T, Steegborn, C, Huber, R, Kessler, D. | | Deposit date: | 2000-03-14 | | Release date: | 2000-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the cystine C-S lyase from Synechocystis: stabilization of cysteine persulfide for FeS cluster biosynthesis.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1YNR
 
 | | Crystal structure of the cytochrome c-552 from Hydrogenobacter thermophilus at 2.0 resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cytochrome c-552, HEME C, ... | | Authors: | Travaglini-Allocatelli, C, Gianni, S, Dubey, V.K, Borgia, A, Di Matteo, A, Bonivento, D, Cutruzzola, F, Bren, K.L, Brunori, M. | | Deposit date: | 2005-01-25 | | Release date: | 2005-05-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Obligatory Intermediate in the Folding Pathway of Cytochrome c552 from Hydrogenobacter thermophilus
J.Biol.Chem., 280, 2005
|
|
1KOK
 
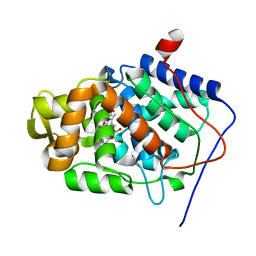 | | Crystal Structure of Mesopone Cytochrome c Peroxidase (MpCcP) | | Descriptor: | Cytochrome c Peroxidase, FE(III)-(4-MESOPORPHYRINONE) | | Authors: | Bhaskar, B, Immoos, C.E, Cohen, M.S, Barrows, T.P, Farmer, P.J, Poulos, T.L. | | Deposit date: | 2001-12-20 | | Release date: | 2002-10-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mesopone cytochrome c peroxidase: functional model of heme oxygenated oxidases.
J.Inorg.Biochem., 91, 2002
|
|
246D
 
 | | STRUCTURE OF THE PURINE-PYRIMIDINE ALTERNATING RNA DOUBLE HELIX, R(GUAUAUA)D(C) , WITH A 3'-TERMINAL DEOXY RESIDUE | | Descriptor: | DNA/RNA (5'-R(*GP*UP*AP*UP*AP*UP*AP*)-D(*C)-3'), SODIUM ION | | Authors: | Wahl, M.C, Ban, C, Sekharudu, C, Ramakrishnan, B, Sundaralingam, M. | | Deposit date: | 1996-01-25 | | Release date: | 1996-08-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the purine-pyrimidine alternating RNA double helix, r(GUAUAUA)d(C), with a 3'-terminal deoxy residue.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
