1PB9
 
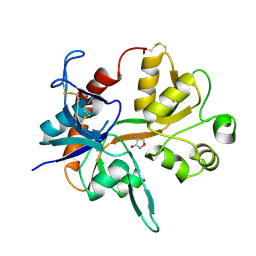 | |
1PBQ
 
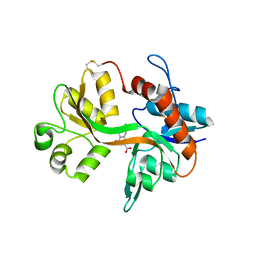 | |
1P1N
 
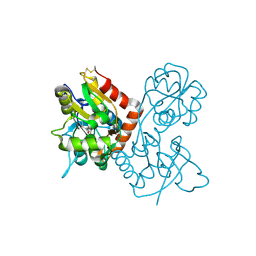 | |
1SYH
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND-BINDING CORE (S1S2J) IN COMPLEX WITH (S)-CPW399 AT 1.85 A RESOLUTION. | | Descriptor: | (S)-2-AMINO-3-(1,3,5,7-PENTAHYDRO-2,4-DIOXO-CYCLOPENTA[E]PYRIMIDIN-1-YL) PROIONIC ACID, Glutamate receptor 2 | | Authors: | Frandsen, A, Pickering, D.S, Vestergaard, B, Kasper, C, Nielsen, B.B, Greenwood, J.R, Campiani, G, Gajhede, M, Schousboe, A, Kastrup, J.S. | | Deposit date: | 2004-04-01 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tyr702 Is an Important Determinant of Agonist Binding and Domain Closure of the Ligand-Binding Core of GluR2.
Mol.Pharmacol., 67, 2005
|
|
4MF3
 
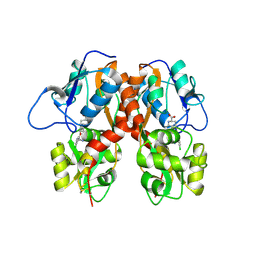 | | Crystal Structure of Human GRIK1 complexed with a 6-(tetrazolyl)aryl decahydroisoquinoline antagonist | | Descriptor: | (3S,4aS,6S,8aR)-6-[3-chloro-2-(1H-tetrazol-5-yl)phenoxy]decahydroisoquinoline-3-carboxylic acid, Glutamate receptor ionotropic, kainate 1 | | Authors: | Martinez-Perez, J.A, Iyengar, S, Shannon, H.E, Bleakman, D, Alt, A, Clawson, D.K, Arnold, B.M, Bell, M.G, Bleisch, T.J, Castano, A.M, Del Prado, M, Dominguez, E, Escribano, A.M, Filla, S.A, Ho, K.H, Hudziak, K.J, Jones, C.K, Katofiasc, M.A, Mateo, A, Mathes, B.M, Mattiuz, E.L, Ogden, A.M.L, Phebus, L.A, Simmons, R.M.A, Stack, D.R, Stratford, R.E, Winter, M.A, Wu, Z, Ornstein, P.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-07 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | GluK1 antagonists from 6-(tetrazolyl)phenyl decahydroisoquinoline derivatives: in vitro profile and in vivo analgesic efficacy.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7LDE
 
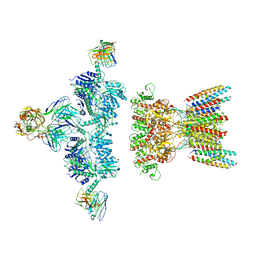 | | native AMPA receptor | | Descriptor: | 11B8 scFv, 15F1 Fab heavy chain, 15F1 Fab light chain, ... | | Authors: | Yu, J, Rao, P, Gouaux, E. | | Deposit date: | 2021-01-13 | | Release date: | 2021-05-12 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Hippocampal AMPA receptor assemblies and mechanism of allosteric inhibition.
Nature, 594, 2021
|
|
7LEP
 
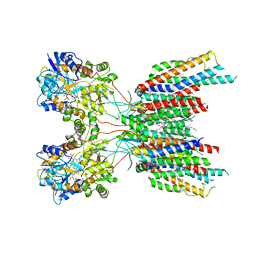 | | The composite LBD-TMD structure combined from all hippocampal AMPAR subtypes at 3.25 Angstrom resolution | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 6-[2-chloro-6-(trifluoromethoxy)phenyl]-1H-benzimidazol-2-ol, DECANE, ... | | Authors: | Yu, J, Rao, P, Gouaux, E. | | Deposit date: | 2021-01-14 | | Release date: | 2021-05-12 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Hippocampal AMPA receptor assemblies and mechanism of allosteric inhibition.
Nature, 594, 2021
|
|
7LDD
 
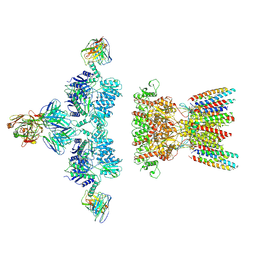 | | native AMPA receptor | | Descriptor: | 11B8 scFv, 15F1 Fab heavy chain, 15F1 Fab light chain, ... | | Authors: | Yu, J, Rao, P, Gouaux, E. | | Deposit date: | 2021-01-13 | | Release date: | 2021-05-12 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Hippocampal AMPA receptor assemblies and mechanism of allosteric inhibition.
Nature, 594, 2021
|
|
4LZ7
 
 | |
4MH5
 
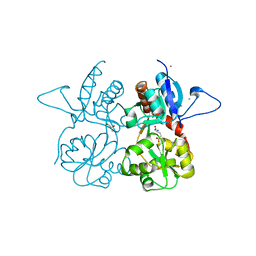 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with (S)-glutamate | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-08-29 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Binding site and interlobe interactions of the ionotropic glutamate receptor GluK3 ligand binding domain revealed by high resolution crystal structure in complex with (S)-glutamate.
J.Struct.Biol., 176, 2011
|
|
7LZI
 
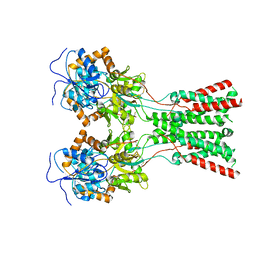 | |
7LZ2
 
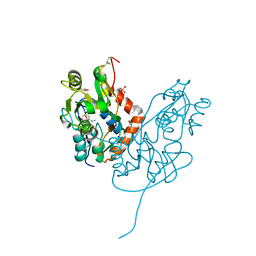 | | Structure of glutamate receptor-like channel GLR3.4 ligand-binding domain in complex with methionine | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gangwar, S.P, Green, M.N, Sobolevsky, A.I. | | Deposit date: | 2021-03-08 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Arabidopsis thaliana glutamate receptor-like channel GLR3.4.
Mol.Cell, 81, 2021
|
|
7LZH
 
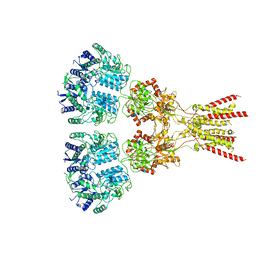 | | Structure of the glutamate receptor-like channel AtGLR3.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Gangwar, S.P, Green, M.N, Sobolevsky, A.I. | | Deposit date: | 2021-03-09 | | Release date: | 2021-07-28 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structure of the Arabidopsis thaliana glutamate receptor-like channel GLR3.4.
Mol.Cell, 81, 2021
|
|
7LZ0
 
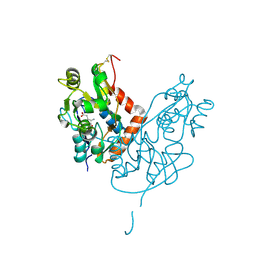 | | Structure of glutamate receptor-like channel GLR3.4 ligand-binding domain in complex with glutamate | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Gangwar, S.P, Green, M.N, Sobolevsky, A.I. | | Deposit date: | 2021-03-08 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure of the Arabidopsis thaliana glutamate receptor-like channel GLR3.4.
Mol.Cell, 81, 2021
|
|
7LZ1
 
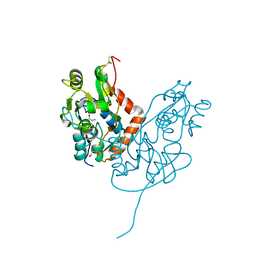 | | Structure of glutamate receptor-like channel GLR3.4 ligand-binding domain in complex with serine | | Descriptor: | GLYCEROL, Glutamate receptor 3.4, SERINE, ... | | Authors: | Gangwar, S.P, Green, M.N, Sobolevsky, A.I. | | Deposit date: | 2021-03-08 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure of the Arabidopsis thaliana glutamate receptor-like channel GLR3.4.
Mol.Cell, 81, 2021
|
|
2PBW
 
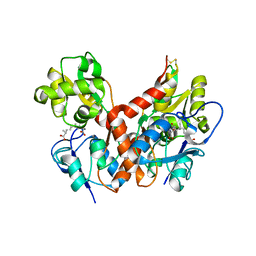 | | Crystal Structure of the Ligand-Binding Core of iGluR5 in Complex with the Partial agonist Domoic Acid at 2.5 A Resolution | | Descriptor: | (2S,3S,4S)-2-CARBOXY-4-[(1Z,3E,5R)-5-CARBOXY-1-METHYL-1,3-HEXADIENYL]-3-PYRROLIDINEACETIC ACID, Glutamate receptor, ionotropic kainate 1 | | Authors: | Hald, H, Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-03-29 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Partial agonism and antagonism of the ionotropic glutamate receptor iGLuR5: structures of the ligand-binding core in complex with domoic acid and 2-amino-3-[5-tert-butyl-3-(phosphonomethoxy)-4-isoxazolyl]propionic acid.
J.Biol.Chem., 282, 2007
|
|
2P2A
 
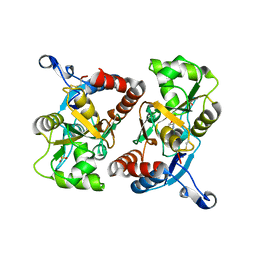 | | X-ray structure of the GluR2 ligand binding core (S1S2J) in complex with 2-Bn-tet-AMPA at 2.26A resolution | | Descriptor: | 2-AMINO-3-[3-HYDROXY-5-(2-BENZYL-2H-5-TETRAZOLYL)-4-ISOXAZOLYL]-PROPIONIC ACID, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Frydenvang, K, Kastrup, J.S, Gajhede, M. | | Deposit date: | 2007-03-07 | | Release date: | 2007-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | A tetrazolyl-substituted subtype-selective AMPA receptor agonist.
J.Med.Chem., 50, 2007
|
|
2QS4
 
 | | Crystal structure of the GluR5 ligand binding core dimer in complex with LY466195 at 1.58 Angstroms resolution | | Descriptor: | (3S,4aR,6S,8aR)-6-{[(2S)-2-carboxy-4,4-difluoropyrrolidin-1-yl]methyl}decahydroisoquinoline-3-carboxylic acid, AMMONIUM ION, GLYCEROL, ... | | Authors: | Alushin, G.M, Jane, D.E, Mayer, M.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
Neuropharmacology, 60, 2011
|
|
4NF6
 
 | | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with glycine and PPDA | | Descriptor: | (2S,3R)-1-(phenanthren-2-ylcarbonyl)piperazine-2,3-dicarboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Jespersen, A, Tajima, N, Furukawa, H. | | Deposit date: | 2013-10-30 | | Release date: | 2014-03-12 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into Competitive Antagonism in NMDA Receptors.
Neuron, 81, 2014
|
|
4NF4
 
 | | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with DCKA and glutamate | | Descriptor: | 4-hydroxy-5,7-dimethylquinoline-2-carboxylic acid, GLUTAMIC ACID, Glutamate receptor ionotropic, ... | | Authors: | Annie, J, Tajima, N, Furukawa, H. | | Deposit date: | 2013-10-30 | | Release date: | 2014-03-12 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into Competitive Antagonism in NMDA Receptors.
Neuron, 81, 2014
|
|
2RCA
 
 | |
2RC9
 
 | |
4N07
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and BPAM-344 at 1.87 A resolution | | Descriptor: | 4-cyclopropyl-7-fluoro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, CACODYLATE ION, ... | | Authors: | Noerholm, A.B, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2013-10-01 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Synthesis, pharmacological and structural characterization, and thermodynamic aspects of GluA2-positive allosteric modulators with a 3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide scaffold.
J.Med.Chem., 56, 2013
|
|
2RC8
 
 | |
4NWD
 
 | | Crystal structure of the kainate receptor GluK3 ligand-binding domain in complex with the agonist (2S,4R)-4-(3-Methylamino-3-oxopropyl)glutamic acid at 2.6 A resolution | | Descriptor: | (4R)-4-[3-(methylamino)-3-oxopropyl]-L-glutamic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | Authors: | Venskutonyte, R, Larsen, A.P, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Recognition of Two 2,4-syn-Functionalized (S)-Glutamate Analogues by the Kainate Receptor GluK3 Ligand Binding Domain.
Chemmedchem, 9, 2014
|
|
