8V15
 
 | | Human SIRT3 bound to p53-AMC peptide, Carba-NAD, and Honokiol | | Descriptor: | (1P)-3',5-di(prop-2-en-1-yl)[1,1'-biphenyl]-2,4'-diol, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, GLN-PRO-LYS-FDL, ... | | Authors: | Chakrabarti, R, Ghosh, A, Guan, X, Upadhyay, A, Dumpati, R.K, Munshi, S, Roy, S, Chall, S, Rahnamoun, A, Reverdy, C, Errasti, G, Delacroix, T. | | Deposit date: | 2023-11-19 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computationally Driven Discovery and Characterization of SIRT3 Activating Compounds that Fully Recover Catalytic Activity under NAD+ Depletion
biorxiv, 2023
|
|
8V2N
 
 | | Human SIRT3 co-crystallized with ligands, including p53-AMC peptide and Carba-NAD | | Descriptor: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, GLN-PRO-LYS-FDL, NAD-dependent protein deacetylase sirtuin-3, ... | | Authors: | Chakrabarti, R, Ghosh, A, Guan, X, Upadhyay, A, Dumpati, R.K, Munshi, S, Roy, S, Chall, S, Rahnamoun, A, Reverdy, C, Errasti, G, Delacroix, T. | | Deposit date: | 2023-11-23 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Computationally Driven Discovery and Characterization of SIRT3 Activating Compounds that Fully Recover Catalytic Activity under NAD+ Depletion
biorxiv, 2023
|
|
8V4O
 
 | |
7MXG
 
 | | PRMT5(M420T mutant):MEP50 complexed with inhibitor PF-06855800 | | Descriptor: | 7-[(5R)-5-C-(4-chloro-3-fluorophenyl)-beta-D-ribofuranosyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | McTigue, M, Deng, Y.L, Liu, W, Brooun, A. | | Deposit date: | 2021-05-19 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | SAM-Competitive PRMT5 Inhibitor PF-06939999 Demonstrates Antitumor Activity in Splicing Dysregulated NSCLC with Decreased Liability of Drug Resistance.
Mol.Cancer Ther., 21, 2022
|
|
6K35
 
 | | Crystal structure of GH20 exo beta-N-acetylglucosaminidase from Vibrio harveyi in complex with NAG-thiazoline | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Beta-N-acetylglucosaminidase Nag2 | | Authors: | Meekrathok, P, Stubbs, K.A, Bulmer, D.M, van den Berg, B, Suginta, W. | | Deposit date: | 2019-05-16 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | NAG-thiazoline is a potent inhibitor of the Vibrio campbellii GH20 beta-N-Acetylglucosaminidase.
Febs J., 287, 2020
|
|
8USH
 
 | |
7NFT
 
 | |
8UZK
 
 | |
8USF
 
 | |
8USG
 
 | |
6K0P
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase D1045A in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.424 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
7NFR
 
 | | Fujian capmidlink domain in complex with Nb8194 | | Descriptor: | GLYCEROL, NB8194, Polymerase basic protein 2, ... | | Authors: | Keown, J.R, Grimes, J.M, Fodor, E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
6BPE
 
 | | Plasmodium vivax reticulocyte binding protein 2b (PvRBP2b) bound to monoclonal antibody 6H1 | | Descriptor: | Monoclonal antibody 6H1 Fab heavy chain, Monoclonal antibody 6H1 Fab light chain, Reticulocyte binding protein 2, ... | | Authors: | Gruszczyk, J, Chan, L.J, Tham, W.H. | | Deposit date: | 2017-11-22 | | Release date: | 2018-06-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Cryo-EM structure of an essential Plasmodium vivax invasion complex.
Nature, 559, 2018
|
|
8V4A
 
 | | Proteus vulgaris tryptophan indole-lyase complexed with L-ethionine | | Descriptor: | (2E)-4-(ethylsulfanyl)-2-{[(Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methyl]imino}butanoic acid, (E)-S-ethyl-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-homocysteine, DIMETHYL SULFOXIDE, ... | | Authors: | Phillips, R.S. | | Deposit date: | 2023-11-28 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Proteus vulgaris tryptophan indole-lyase complexed with L-ethionine
To Be Published
|
|
8V5G
 
 | |
6C8Z
 
 | | Last common ancestor of ADP-dependent phosphofructokinases from Methanosarcinales | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADP-dependent phosphofructokinase, MAGNESIUM ION, ... | | Authors: | Castro-Fernandez, V, Gonzalez-Ordenes, F, Munoz, S, Fuentes, N, Leonardo, D, Fuentealba, M, Herrera-Morande, A, Maturana, P, Villalobos, P, Garratt, R. | | Deposit date: | 2018-01-25 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | ADP-Dependent Kinases From the Archaeal OrderMethanosarcinalesAdapt to Salt by a Non-canonical Evolutionarily Conserved Strategy.
Front Microbiol, 9, 2018
|
|
7NFQ
 
 | | Fujian capmidlink domain in complex with Nb8193 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Nb8193, ... | | Authors: | Keown, J.R, Grimes, J.M, Fodor, E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
8V4J
 
 | | Phosphoheptose isomerase GMHA from Burkholderia pseudomallei bound to inhibitor Mut148233 | | Descriptor: | 1-deoxy-1-[formyl(hydroxy)amino]-5-O-phosphono-D-ribitol, CHLORIDE ION, Phosphoheptose isomerase, ... | | Authors: | Junop, M.S, Brown, C, Szabla, R. | | Deposit date: | 2023-11-29 | | Release date: | 2023-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Potentiating Activity of GmhA Inhibitors on Gram-Negative Bacteria.
J.Med.Chem., 67, 2024
|
|
7NIZ
 
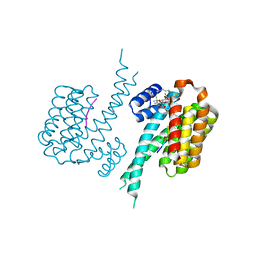 | | Human 14-3-3 sigma in complex with human Estrogen Receptor alpha peptide and ligands Fusicoccin-A and WR-1065 | | Descriptor: | 14-3-3 protein sigma, Estrogen receptor, FUSICOCCIN, ... | | Authors: | Roversi, P, Falcicchio, M, Doveston, R. | | Deposit date: | 2021-02-14 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Cooperative stabilisation of 14-3-3 sigma protein-protein interactions via covalent protein modification.
Chem Sci, 12, 2021
|
|
8UK9
 
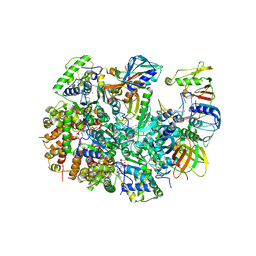 | | Structure of T4 Bacteriophage clamp loader mutant D110C bound to the T4 clamp, primer-template DNA, and ATP analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, DNA primer, ... | | Authors: | Marcus, K, Ghaffari-Kashani, S, Gee, C.L. | | Deposit date: | 2023-10-12 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8UH7
 
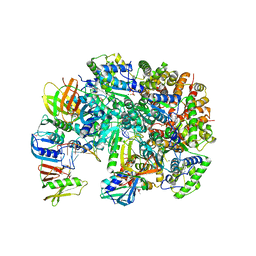 | | Structure of T4 Bacteriophage clamp loader bound to the T4 clamp, primer-template DNA, and ATP analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Primer DNA strand, ... | | Authors: | Gee, C.L, Marcus, K, Kelch, B.A, Makino, D.L. | | Deposit date: | 2023-10-07 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.628 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6C9J
 
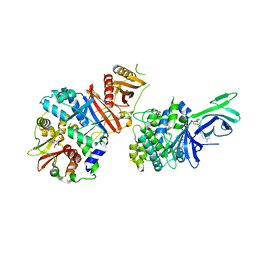 | | AMP-activated protein kinase bound to pharmacological activator R734 | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Zhou, X.E, Novick, S, Shaw, S.J, Li, Y, Brunzelle, J.S, Hitoshi, Y, Griffin, P.R, Xu, H.E, Melcher, K. | | Deposit date: | 2018-01-26 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structures of AMP-activated protein kinase bound to novel pharmacological activators in phosphorylated, non-phosphorylated, and nucleotide-free states.
J. Biol. Chem., 294, 2019
|
|
6K8M
 
 | |
6BW1
 
 | | Hendra virus W protein C-terminus in complex with Importin alpha 1 | | Descriptor: | Importin subunit alpha-1, Protein W | | Authors: | Tsimbalyuk, S, Smith, K.M, Edwards, M.R, Aragao, D, Cross, E.M, Basler, C.F, Forwood, J.K. | | Deposit date: | 2017-12-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses.
Nat Commun, 9, 2018
|
|
6KAK
 
 | | Crystal structure of FKRP in complex with Mg ion | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fukutin-related protein, MAGNESIUM ION, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2019-06-23 | | Release date: | 2020-01-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.056 Å) | | Cite: | Crystal structures of fukutin-related protein (FKRP), a ribitol-phosphate transferase related to muscular dystrophy.
Nat Commun, 11, 2020
|
|
